Streptococcus pneumoniae TIGR4 Flavodoxin: Structural and Biophysical Characterization of a Novel Drug Target
- PMID: 27649488
- PMCID: PMC5029806
- DOI: 10.1371/journal.pone.0161020
Streptococcus pneumoniae TIGR4 Flavodoxin: Structural and Biophysical Characterization of a Novel Drug Target
Abstract
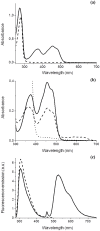
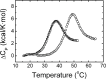
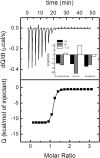
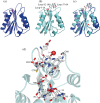
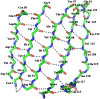
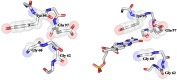
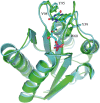
Streptococcus pneumoniae (Sp) strain TIGR4 is a virulent, encapsulated serotype that causes bacteremia, otitis media, meningitis and pneumonia. Increased bacterial resistance and limited efficacy of the available vaccine to some serotypes complicate the treatment of diseases associated to this microorganism. Flavodoxins are bacterial proteins involved in several important metabolic pathways. The Sp flavodoxin (Spfld) gene was recently reported to be essential for the establishment of meningitis in a rat model, which makes SpFld a potential drug target. To facilitate future pharmacological studies, we have cloned and expressed SpFld in E. coli and we have performed an extensive structural and biochemical characterization of both the apo form and its active complex with the FMN cofactor. SpFld is a short-chain flavodoxin containing 146 residues. Unlike the well-characterized long-chain apoflavodoxins, the Sp apoprotein displays a simple two-state thermal unfolding equilibrium and binds FMN with moderate affinity. The X-ray structures of the apo and holo forms of SpFld differ at the FMN binding site, where substantial rearrangement of residues at the 91-100 loop occurs to permit cofactor binding. This work will set up the basis for future studies aiming at discovering new potential drugs to treat S. pneumoniae diseases through the inhibition of SpFld.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Thermal unfolding of Apo and Holo Desulfovibrio desulfuricans flavodoxin: cofactor stabilizes folded and intermediate states.Biochemistry. 2004 Oct 12;43(40):12855-64. doi: 10.1021/bi048944e. Biochemistry. 2004. PMID: 15461458
-
Expression and characterization of the two flavodoxin proteins of Bacillus subtilis, YkuN and YkuP: biophysical properties and interactions with cytochrome P450 BioI.Biochemistry. 2004 Oct 5;43(39):12390-409. doi: 10.1021/bi049131t. Biochemistry. 2004. PMID: 15449930
-
How FMN binds to anabaena apoflavodoxin: a hydrophobic encounter at an open binding site.J Biol Chem. 2003 Jun 27;278(26):24053-61. doi: 10.1074/jbc.M301049200. Epub 2003 Apr 7. J Biol Chem. 2003. PMID: 12682068
-
The midpoint potentials for the oxidized-semiquinone couple for Gly57 mutants of the Clostridium beijerinckii flavodoxin correlate with changes in the hydrogen-bonding interaction with the proton on N(5) of the reduced flavin mononucleotide cofactor as measured by NMR chemical shift temperature dependencies.Biochemistry. 1999 Jun 1;38(22):7168-76. doi: 10.1021/bi982203u. Biochemistry. 1999. PMID: 10353827
-
Flavodoxins: sequence, folding, binding, function and beyond.Cell Mol Life Sci. 2006 Apr;63(7-8):855-64. doi: 10.1007/s00018-005-5514-4. Cell Mol Life Sci. 2006. PMID: 16465441 Free PMC article. Review.
Cited by
-
Gallic acid based green corrosion inhibitor for mild steel in 1 M HCl electrochemical and microbial assessment with theoretical validation.Sci Rep. 2025 Apr 30;15(1):15156. doi: 10.1038/s41598-025-97647-3. Sci Rep. 2025. PMID: 40307297 Free PMC article.
-
Synthesis of Novel Nano-Sulfonamide Metal-Based Corrosion Inhibitor Surfactants.Materials (Basel). 2022 Feb 1;15(3):1146. doi: 10.3390/ma15031146. Materials (Basel). 2022. PMID: 35161090 Free PMC article.
-
Characterization of Fusobacterium varium Fv113-g1 isolated from a patient with ulcerative colitis based on complete genome sequence and transcriptome analysis.PLoS One. 2017 Dec 7;12(12):e0189319. doi: 10.1371/journal.pone.0189319. eCollection 2017. PLoS One. 2017. PMID: 29216329 Free PMC article.
-
L-Thyroxine and L-thyroxine-based antimicrobials against Streptococcus pneumoniae and other Gram-positive bacteria.Heliyon. 2024 Mar 22;10(7):e27982. doi: 10.1016/j.heliyon.2024.e27982. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38689973 Free PMC article.
-
In silico Prediction of New Drug Candidates Against the Multidrug-Resistant and Potentially Zoonotic Fish Pathogen Serotype III Streptococcus agalactiae.Front Genet. 2020 Aug 28;11:1024. doi: 10.3389/fgene.2020.01024. eCollection 2020. Front Genet. 2020. PMID: 33005185 Free PMC article.
References
-
- Lawson RJ, von Wachenfeldt C, Haq I, Perkins J, Munro AW. Expression and characterization of the two flavodoxin proteins of Bacillus subtilis, YkuN and YkuP: biophysical properties and interactions with cytochrome P450 BioI. Biochemistry, 2004. 43(39): p. 12390–12409. - PubMed
-
- Martin M, Turco JH, Zegans ME, Facklam RR, Sodha S, Elliott JA, et al. An Outbreak of Conjunctivitis Due to Atypical Streptococcus pneumoniae. New England Journal of Medicine, 2003. 348(12): p. 1112–1121. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

