Quantitative Analysis of the Microtubule Interaction of Rabies Virus P3 Protein: Roles in Immune Evasion and Pathogenesis
- PMID: 27649849
- PMCID: PMC5030706
- DOI: 10.1038/srep33493
Quantitative Analysis of the Microtubule Interaction of Rabies Virus P3 Protein: Roles in Immune Evasion and Pathogenesis
Abstract
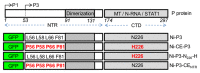
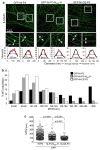
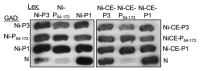
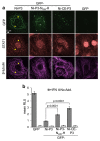
Although microtubules (MTs) are known to have important roles in intracellular transport of many viruses, a number of reports suggest that specific viral MT-associated proteins (MAPs) target MTs to subvert distinct MT-dependent cellular processes. The precise functional importance of these interactions and their roles in pathogenesis, however, remain largely unresolved. To assess the association with disease of the rabies virus (RABV) MAP, P3, we quantitatively compared the phenotypes of P3 from a pathogenic RABV strain, Nishigahara (Ni) and a non-pathogenic Ni-derivative strain, Ni-CE. Using confocal/live-cell imaging and dSTORM super-resolution microscopy to quantify protein interactions with the MT network and with individual MT filaments, we found that the interaction by Ni-CE-P3 is significantly impaired compared with Ni-P3. This correlated with an impaired capacity to effect association of the transcription factor STAT1 with MTs and to antagonize interferon (IFN)/STAT1-dependent antiviral signaling. Importantly, we identified a single mutation in Ni-CE-P3 that is sufficient to inhibit MT-association and IFN-antagonist function of Ni-P3, and showed that this mutation alone attenuates the pathogenicity of RABV. These data provide evidence that the viral protein-MT interface has important roles in pathogenesis, suggesting that this interface could provide targets for vaccine/antiviral drug development.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

