A conserved influenza A virus nucleoprotein code controls specific viral genome packaging
- PMID: 27650413
- PMCID: PMC5035998
- DOI: 10.1038/ncomms12861
A conserved influenza A virus nucleoprotein code controls specific viral genome packaging
Abstract
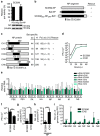
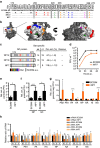
Packaging of the eight genomic RNA segments of influenza A viruses (IAV) into viral particles is coordinated by segment-specific packaging sequences. How the packaging signals regulate the specific incorporation of each RNA segment into virions and whether other viral or host factors are involved in this process is unknown. Here, we show that distinct amino acids of the viral nucleoprotein (NP) are required for packaging of specific RNA segments. This was determined by studying the NP of a bat influenza A-like virus, HL17NL10, in the context of a conventional IAV (SC35M). Replacement of conserved SC35M NP residues by those of HL17NL10 NP resulted in RNA packaging defective IAV. Surprisingly, substitution of these conserved SC35M amino acids with HL17NL10 NP residues led to IAV with altered packaging efficiencies for specific subsets of RNA segments. This suggests that NP harbours an amino acid code that dictates genome packaging into infectious virions.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

