MBD3 expression and DNA binding patterns are altered in a rat model of temporal lobe epilepsy
- PMID: 27650712
- PMCID: PMC5030630
- DOI: 10.1038/srep33736
MBD3 expression and DNA binding patterns are altered in a rat model of temporal lobe epilepsy
Abstract
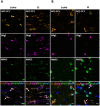
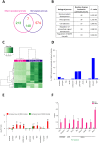
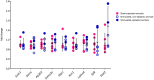
The aim of the present study was to examine involvement of MBD3 (methyl-CpG-binding domain protein 3), a protein involved in reading DNA methylation patterns, in epileptogenesis and epilepsy. We used a well-characterized rat model of temporal lobe epilepsy that is triggered by status epilepticus, evoked by electrical stimulation of the amygdala. Stimulated and sham-operated animals were sacrificed 14 days after stimulation. We found that MBD3 transcript was present in neurons, oligodendrocytes, and astrocytes in both control and epileptic animals. We detected the nuclear localization of MBD3 protein in neurons, mature oligodendrocytes, and a subpopulation of astrocytes but not in microglia. Amygdala stimulation significantly increased the level of MBD3 immunofluorescence. Immunoprecipitation followed by mass spectrometry and Western blot revealed that MBD3 in the adult brain assembles the NuRD complex, which also contains MTA2, HDAC2, and GATAD2B. Using chromatin immunoprecipitation combined with deep sequencing, we observed differences in the occupancy of DNA regions by MBD3 protein between control and stimulated animals. This was not followed by subsequent changes in the mRNA expression levels of selected MBD3 targets. Our data demonstrate for the first time alterations in the MBD3 expression and DNA occupancy in the experimental model of epilepsy.
Figures





Similar articles
-
Methyl-CpG-Binding Domain Protein 3 Promotes Seizures by Recruiting Methyltransferase DNMT1 to Enhance TREM2 Methylation.Neurochem Res. 2021 Sep;46(9):2451-2462. doi: 10.1007/s11064-021-03371-5. Epub 2021 Jun 25. Neurochem Res. 2021. PMID: 34173118
-
Epileptogenesis-Associated Alterations of Heat Shock Protein 70 in a Rat Post-Status Epilepticus Model.Neuroscience. 2019 Sep 1;415:44-58. doi: 10.1016/j.neuroscience.2019.06.031. Epub 2019 Jul 15. Neuroscience. 2019. PMID: 31319099
-
Progression of neuronal damage after status epilepticus and during spontaneous seizures in a rat model of temporal lobe epilepsy.Prog Brain Res. 2002;135:67-83. doi: 10.1016/S0079-6123(02)35008-8. Prog Brain Res. 2002. PMID: 12143371
-
A new model of chronic temporal lobe epilepsy induced by electrical stimulation of the amygdala in rat.Epilepsy Res. 2000 Feb;38(2-3):177-205. doi: 10.1016/s0920-1211(99)00088-1. Epilepsy Res. 2000. PMID: 10642046
-
Amygdala damage in experimental and human temporal lobe epilepsy.Epilepsy Res. 1998 Sep;32(1-2):233-53. doi: 10.1016/s0920-1211(98)00055-2. Epilepsy Res. 1998. PMID: 9761324 Review.
Cited by
-
Behavioral characteristics as potential biomarkers of the development and phenotype of epilepsy in a rat model of temporal lobe epilepsy.Sci Rep. 2021 Apr 21;11(1):8665. doi: 10.1038/s41598-021-88088-9. Sci Rep. 2021. PMID: 33883658 Free PMC article.
-
Uncovering the Molecular Interactions Underlying MBD2 and MBD3 Phase Separation.J Phys Chem B. 2025 Jun 12;129(23):5728-5743. doi: 10.1021/acs.jpcb.5c02741. Epub 2025 May 11. J Phys Chem B. 2025. PMID: 40350613 Free PMC article.
-
Deconvolution of Transcriptional Networks in Post-Traumatic Stress Disorder Uncovers Master Regulators Driving Innate Immune System Function.Sci Rep. 2017 Nov 3;7(1):14486. doi: 10.1038/s41598-017-15221-y. Sci Rep. 2017. PMID: 29101382 Free PMC article.
References
-
- Porter R. J. In A textbook of epilepsy (eds Laidlaw J. et al.) 1–22 (Churchil Livingstone, 1993).
-
- Hauser W. A. In Epilepsy: a Comprehensive Textbook (eds Engel J. Jr & Pedley T. A.) 47–57 (Lippincott-Raven Publishers, 1997).
-
- Pitkanen A. & Sutula T. P. Is epilepsy a progressive disorder? Prospects for new therapeutic approaches in temporal-lobe epilepsy. Lancet Neurol 1, 173–181 (2002). - PubMed
-
- Pitkanen A. & Lukasiuk K. Mechanisms of epileptogenesis and potential treatment targets. Lancet Neurol 10, 173–186 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

