Nutritional Stress Induced by Tryptophan-Degrading Enzymes Results in ATF4-Dependent Reprogramming of the Amino Acid Transporter Profile in Tumor Cells
- PMID: 27651314
- PMCID: PMC5096689
- DOI: 10.1158/0008-5472.CAN-15-3502
Nutritional Stress Induced by Tryptophan-Degrading Enzymes Results in ATF4-Dependent Reprogramming of the Amino Acid Transporter Profile in Tumor Cells
Abstract
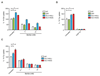
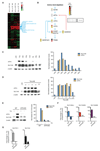
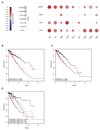
Tryptophan degradation is an immune escape strategy shared by many tumors. However, cancer cells' compensatory mechanisms remain unclear. We demonstrate here that a shortage of tryptophan caused by expression of indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO) resulted in ATF4-dependent upregulation of several amino acid transporters, including SLC1A5 and its truncated isoforms, which in turn enhanced tryptophan and glutamine uptake. Importantly, SLC1A5 failed to be upregulated in resting human T cells kept under low tryptophan conditions but was enhanced upon cognate antigen T-cell receptor engagement. Our results highlight key differences in the ability of tumor and T cells to adapt to tryptophan starvation and provide important insights into the poor prognosis of tumors coexpressing IDO and SLC1A5. Cancer Res; 76(21); 6193-204. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures



References
-
- Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nature Medicine. 2003;9(10):1269–74. - PubMed
-
- Theate I, van Baren N, Pilotte L, Moulin P, Larrieu P, Renauld J-C et al. Extensive Profiling of the Expression of the Indoleamine 2,3-Dioxygenase 1 Protein in Normal and Tumoral Human Tissues. Cancer Immunology Research. 2015;3(2):161–72. - PubMed
-
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478(7368):197–203. - PubMed
-
- Godin-Ethier J, Hanafi L-A, Piccirillo CA, Lapointe R. Indoleamine 2,3-Dioxygenase Expression in Human Cancers: Clinical and Immunologic Perspectives. Clinical Cancer Research. 2011;17(22):6985–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

