Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI
- PMID: 27651330
- PMCID: PMC5263212
- DOI: 10.1007/s11548-016-1483-3
Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI
Abstract
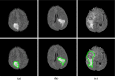
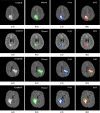
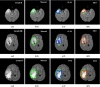
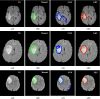
Purpose: We propose a fully automated method for detection and segmentation of the abnormal tissue associated with brain tumour (tumour core and oedema) from Fluid- Attenuated Inversion Recovery (FLAIR) Magnetic Resonance Imaging (MRI).
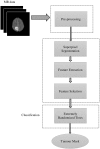
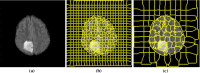
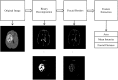
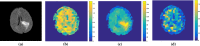
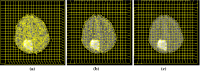
Methods: The method is based on superpixel technique and classification of each superpixel. A number of novel image features including intensity-based, Gabor textons, fractal analysis and curvatures are calculated from each superpixel within the entire brain area in FLAIR MRI to ensure a robust classification. Extremely randomized trees (ERT) classifier is compared with support vector machine (SVM) to classify each superpixel into tumour and non-tumour.
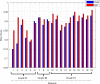
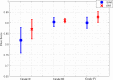
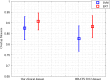
Results: The proposed method is evaluated on two datasets: (1) Our own clinical dataset: 19 MRI FLAIR images of patients with gliomas of grade II to IV, and (2) BRATS 2012 dataset: 30 FLAIR images with 10 low-grade and 20 high-grade gliomas. The experimental results demonstrate the high detection and segmentation performance of the proposed method using ERT classifier. For our own cohort, the average detection sensitivity, balanced error rate and the Dice overlap measure for the segmented tumour against the ground truth are 89.48 %, 6 % and 0.91, respectively, while, for the BRATS dataset, the corresponding evaluation results are 88.09 %, 6 % and 0.88, respectively.
Conclusions: This provides a close match to expert delineation across all grades of glioma, leading to a faster and more reproducible method of brain tumour detection and delineation to aid patient management.
Keywords: Brain tumour segmentation; Extremely randomized trees; Feature selection; Magnetic resonance imaging; Superpixels; Textons.
Conflict of interest statement
The authors declare they have no conflict of interest with regard to the work presented. Ethical approval Ethical approval to conduct this study was obtained from Local Committee of St George’s Hospital, London. Informed consent Informed consent was obtained from all individual participants in the study.
Figures




References
-
- Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, DeGroot J, Wick W, Gilbert MR, Lassman AB, Tsien C, Mikkelsen T, Wong ET, Chamberlain MC, Stupp R, Lamborn KR, Vogelbaum MA, van den Bent MJ, Chang SM. Updated response assessment criteria for high-grade gliomas: response assessment in Neuro-Oncology Working Group. JCO. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. - DOI - PubMed
-
- Niyazi M, Brada M, Chalmers AJ, Combs SE, Erridge SC, Fiorentino A, Grosu AL, Lagerwaard FJ, Minniti G, Mirimanoff R-O, Ricardi U, Short SC, Weber DC, Belka C. ESTRO-ACROP guideline “target delineation of glioblastomas.”. Radiother Oncol. 2016;118:35–42. doi: 10.1016/j.radonc.2015.12.003. - DOI - PubMed
-
- Aslian H, Sadeghi M, Mahdavi SR, Babapour Mofrad F, Astarakee M, Khaledi N, Fadavi P. Magnetic resonance imaging-based target volume delineation in radiation therapy treatment planning for brain tumors using localized region-based active contour. Int J Radiat Oncol Biol Phys. 2013;87:195–201. doi: 10.1016/j.ijrobp.2013.04.049. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

