Emergence of a Potent Multidrug Efflux Pump Variant That Enhances Campylobacter Resistance to Multiple Antibiotics
- PMID: 27651364
- PMCID: PMC5030363
- DOI: 10.1128/mBio.01543-16
Emergence of a Potent Multidrug Efflux Pump Variant That Enhances Campylobacter Resistance to Multiple Antibiotics
Abstract
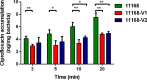
Bacterial antibiotic efflux pumps are key players in antibiotic resistance. Although their role in conferring multidrug resistance is well documented, the emergence of "super" efflux pump variants that enhance bacterial resistance to multiple drugs has not been reported. Here, we describe the emergence of a resistance-enhancing variant (named RE-CmeABC) of the predominant efflux pump CmeABC in Campylobacter, a major zoonotic pathogen whose resistance to antibiotics is considered a serious antibiotic resistance threat in the United States. Compared to the previously characterized CmeABC transporters, RE-CmeABC is much more potent in conferring Campylobacter resistance to antibiotics, which was shown by increased MICs and reduced intracellular accumulation of antibiotics. Structural modeling suggests that sequence variations in the drug-binding pocket of CmeB possibly contribute to the enhanced efflux function. Additionally, RE-CmeABC expands the mutant selection window of ciprofloxacin, enhances the emergence of antibiotic-resistant mutants, and confers exceedingly high-level resistance to fluoroquinolones, an important class of antibiotics for clinical therapy of campylobacteriosis. Furthermore, RE-CmeABC is horizontally transferable, shifts antibiotic MIC distribution among clinical isolates, and is increasingly prevalent in Campylobacter jejuni isolates, suggesting that it confers a fitness advantage under antimicrobial selection. These findings reveal a new mechanism for enhanced multidrug resistance and an effective strategy utilized by bacteria for adaptation to selection from multiple antibiotics.
Importance: Bacterial antibiotic efflux pumps are ubiquitously present in bacterial organisms and protect bacteria from the antibacterial effects of antimicrobials and other toxic compounds by extruding them out of cells. Thus, these efflux transporters represent an important mechanism for antibiotic resistance. In this study, we discovered the emergence and increasing prevalence of a unique efflux pump variant that is much more powerful in the efflux of antibiotics and confers multidrug resistance in Campylobacter, which is a major foodborne pathogen transmitted to humans via the food chain. Unlike other specific resistance determinants that only allow bacteria to resist a particular antimicrobial, the acquisition of a functionally enhanced efflux pump will empower bacteria with simultaneous resistance to multiple classes of antibiotics. These findings reveal a previously undescribed mechanism for enhanced multidrug resistance and open a new direction for us to understand how bacteria adapt to antibiotic treatment.
Copyright © 2016 Yao et al.
Figures


Similar articles
-
Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.Microbiol Spectr. 2023 Aug 17;11(4):e0119723. doi: 10.1128/spectrum.01197-23. Epub 2023 Jun 8. Microbiol Spectr. 2023. PMID: 37289051 Free PMC article.
-
Mutation-based mechanism and evolution of the potent multidrug efflux pump RE-CmeABC in Campylobacter.Proc Natl Acad Sci U S A. 2024 Dec 17;121(51):e2415823121. doi: 10.1073/pnas.2415823121. Epub 2024 Nov 27. Proc Natl Acad Sci U S A. 2024. PMID: 39602248 Free PMC article.
-
Effect of an efflux pump inhibitor on the function of the multidrug efflux pump CmeABC and antimicrobial resistance in Campylobacter.Foodborne Pathog Dis. 2006 Winter;3(4):393-402. doi: 10.1089/fpd.2006.3.393. Foodborne Pathog Dis. 2006. PMID: 17199521
-
Antimicrobial Resistance in Campylobacter spp.Microbiol Spectr. 2018 Apr;6(2):10.1128/microbiolspec.arba-0013-2017. doi: 10.1128/microbiolspec.ARBA-0013-2017. Microbiol Spectr. 2018. PMID: 29623873 Free PMC article. Review.
-
Antibiotic resistance trends and mechanisms in the foodborne pathogen, Campylobacter.Anim Health Res Rev. 2017 Dec;18(2):87-98. doi: 10.1017/S1466252317000135. Epub 2017 Nov 23. Anim Health Res Rev. 2017. PMID: 29166961 Review.
Cited by
-
Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.mBio. 2024 Mar 13;15(3):e0343023. doi: 10.1128/mbio.03430-23. Epub 2024 Feb 7. mBio. 2024. PMID: 38323832 Free PMC article.
-
Genotypic characterization and antimicrobial susceptibility of human Campylobacter jejuni isolates in Southern Spain.Microbiol Spectr. 2024 Oct 3;12(10):e0102824. doi: 10.1128/spectrum.01028-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162511 Free PMC article.
-
Point Deletion or Insertion in CmeR-Box, A2075G Substitution in 23S rRNA, and Presence of erm(B) Are Key Factors of Erythromycin Resistance in Campylobacter jejuni and Campylobacter coli Isolated From Central China.Front Microbiol. 2020 Mar 3;11:203. doi: 10.3389/fmicb.2020.00203. eCollection 2020. Front Microbiol. 2020. PMID: 32194516 Free PMC article.
-
High Prevalence and Predominance of the aph(2″)-If Gene Conferring Aminoglycoside Resistance in Campylobacter.Antimicrob Agents Chemother. 2017 Apr 24;61(5):e00112-17. doi: 10.1128/AAC.00112-17. Print 2017 May. Antimicrob Agents Chemother. 2017. PMID: 28264854 Free PMC article.
-
The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018.EFSA J. 2020 Mar 3;18(3):e06007. doi: 10.2903/j.efsa.2020.6007. eCollection 2020 Mar. EFSA J. 2020. PMID: 32874244 Free PMC article.
References
-
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi:10.1128/AAC.00774-09. - DOI - PMC - PubMed
-
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi:10.1016/S1473-3099(10)70143-2. - DOI - PMC - PubMed
-
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi:10.1016/S1473-3099(15)00424-7. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

