Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases
- PMID: 27651493
- PMCID: PMC5047200
- DOI: 10.1073/pnas.1606004113
Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases
Abstract
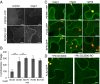
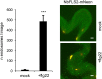
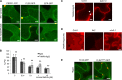
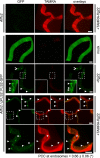
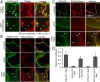
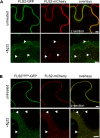
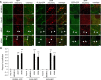
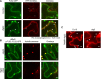
Sensing of potential pathogenic bacteria is of critical importance for immunity. In plants, this involves plasma membrane-resident pattern recognition receptors, one of which is the FLAGELLIN SENSING 2 (FLS2) receptor kinase. Ligand-activated FLS2 receptors are internalized into endosomes. However, the extent to which these spatiotemporal dynamics are generally present among pattern recognition receptors (PRRs) and their regulation remain elusive. Using live-cell imaging, we show that at least three other receptor kinases associated with plant immunity, PEP RECEPTOR 1/2 (PEPR1/2) and EF-TU RECEPTOR (EFR), internalize in a ligand-specific manner. In all cases, endocytosis requires the coreceptor BRI1-ASSOCIATED KINASE 1 (BAK1), and thus depends on receptor activation status. We also show the internalization of liganded FLS2, suggesting the transport of signaling competent receptors. Trafficking of activated PRRs requires clathrin and converges onto the same endosomal vesicles that are also shared with the hormone receptor BRASSINOSTERIOD INSENSITIVE 1 (BRI1). Importantly, clathrin-dependent endocytosis participates in plant defense against bacterial infection involving FLS2-mediated stomatal closure and callose deposition, but is uncoupled from activation of the flagellin-induced oxidative burst and MAP kinase signaling. In conclusion, immunity mediated by pattern recognition receptors depends on clathrin, a critical component for the endocytosis of signaling competent receptors into a common endosomal pathway.
Keywords: EFR; FLS2; PEPR1; clathrin; pattern-triggered immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
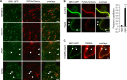
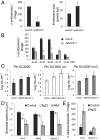
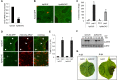
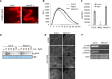

Comment in
- 10745
Similar articles
-
The Arabidopsis malectin-like leucine-rich repeat receptor-like kinase IOS1 associates with the pattern recognition receptors FLS2 and EFR and is critical for priming of pattern-triggered immunity.Plant Cell. 2014 Jul;26(7):3201-19. doi: 10.1105/tpc.114.125682. Epub 2014 Jul 28. Plant Cell. 2014. Retraction in: Plant Cell. 2015 May;27(5):1563. doi: 10.1105/tpc.15.00374. PMID: 25070640 Free PMC article. Retracted.
-
A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence.Nature. 2007 Jul 26;448(7152):497-500. doi: 10.1038/nature05999. Epub 2007 Jul 11. Nature. 2007. PMID: 17625569
-
Sterols regulate endocytic pathways during flg22-induced defense responses in Arabidopsis.Development. 2018 Oct 1;145(19):dev165688. doi: 10.1242/dev.165688. Development. 2018. PMID: 30228101
-
Vesicle trafficking in plant immune responses.Cell Microbiol. 2007 Jan;9(1):1-8. doi: 10.1111/j.1462-5822.2006.00829.x. Epub 2006 Nov 1. Cell Microbiol. 2007. PMID: 17081192 Review.
-
A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors.Annu Rev Plant Biol. 2009;60:379-406. doi: 10.1146/annurev.arplant.57.032905.105346. Annu Rev Plant Biol. 2009. PMID: 19400727 Review.
Cited by
-
Plant elicitor peptide induces endocytosis of plasma membrane proteins in Arabidopsis.Front Plant Sci. 2023 Dec 22;14:1328250. doi: 10.3389/fpls.2023.1328250. eCollection 2023. Front Plant Sci. 2023. PMID: 38186590 Free PMC article.
-
Juxta-membrane S-acylation of plant receptor-like kinases is likely fortuitous and does not necessarily impact upon function.Sci Rep. 2019 Sep 6;9(1):12818. doi: 10.1038/s41598-019-49302-x. Sci Rep. 2019. PMID: 31492958 Free PMC article.
-
Using the CODIT model to explain secondary metabolites of xylem in defence systems of temperate trees against decay fungi.Ann Bot. 2020 Apr 25;125(5):701-720. doi: 10.1093/aob/mcz138. Ann Bot. 2020. PMID: 31420666 Free PMC article. Review.
-
The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses.Prog Lipid Res. 2022 Apr;86:101158. doi: 10.1016/j.plipres.2022.101158. Epub 2022 Feb 5. Prog Lipid Res. 2022. PMID: 35134459 Free PMC article. Review.
-
Nanobody-Dependent Delocalization of Endocytic Machinery in Arabidopsis Root Cells Dampens Their Internalization Capacity.Front Plant Sci. 2021 Mar 19;12:538580. doi: 10.3389/fpls.2021.538580. eCollection 2021. Front Plant Sci. 2021. PMID: 33815429 Free PMC article.
References
-
- Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T. Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol. 2014;20:47–54. - PubMed
-
- Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. - PubMed
-
- Sun Y, et al. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science. 2013;342(6158):624–628. - PubMed
-
- Somssich M, et al. Real-time dynamics of peptide ligand-dependent receptor complex formation in planta. Sci Signal. 2015;8(388):ra76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

