Molecular Pap smear: HPV genotype and DNA methylation of ADCY8, CDH8, and ZNF582 as an integrated biomarker for high-grade cervical cytology
- PMID: 27651839
- PMCID: PMC5022163
- DOI: 10.1186/s13148-016-0263-9
Molecular Pap smear: HPV genotype and DNA methylation of ADCY8, CDH8, and ZNF582 as an integrated biomarker for high-grade cervical cytology
Abstract
Background: The Pap smear has remained the foundation for cervical cancer screening for over 70 years. With advancements in molecular diagnostics, primary high-risk human papillomavirus (hrHPV) screening has recently become an accepted stand-alone or co-test with conventional cytology. However, both diagnostic tests have distinct limitations. The aim of this study was to determine the association between HPV genotypes and cellular epigenetic modifications in three grades of cervical cytology for screening biomarker discovery.
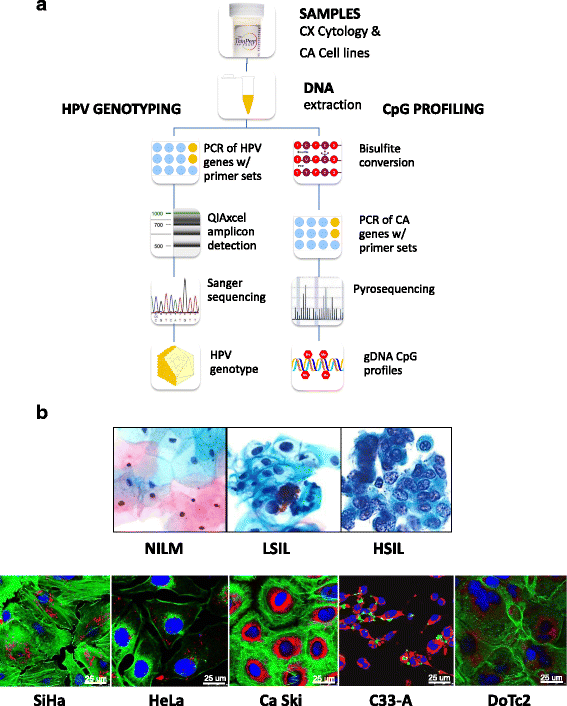
Methods: This prospective, cross-sectional study used residual liquid-based cytology samples for HPV genotyping and epigenetic analysis. Extracted DNA was subjected to parallel polymerase chain reactions using three primer sets (MY09/11, FAP59/64, E6-E7 F/B) for HPV DNA amplification. HPV+ samples were genotyped by DNA sequencing. Promoter methylation of four candidate tumor suppressor genes (adenylate cyclase 8 (ADCY8), cadherin 8, type 2 (CDH8), MGMT, and zinc finger protein 582 (ZNF582)) out of 48 genes screened was quantified by bisulfite-pyrosequencing of genomic DNA. Independent validation of methylation profiles was performed by analyzing data from cervical cancer cell lines and clinical samples from The Cancer Genome Atlas (TCGA).
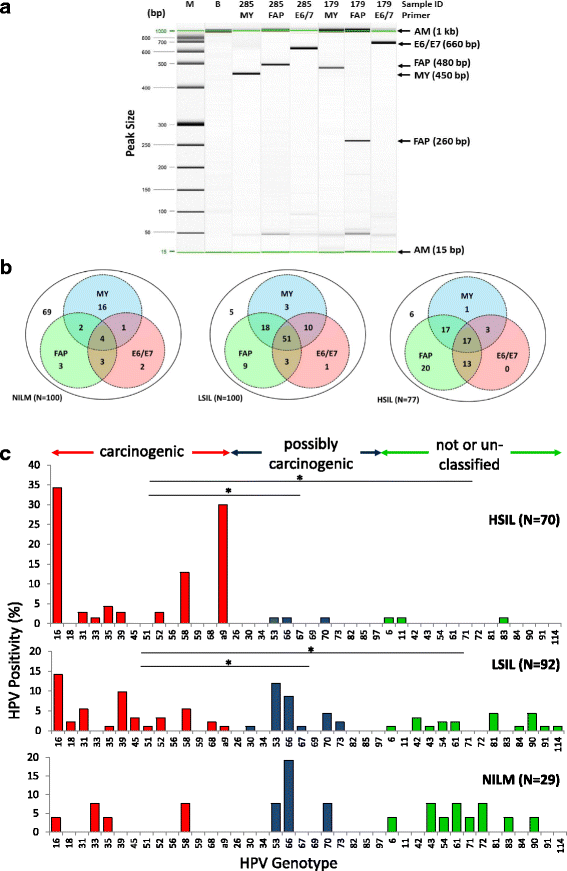
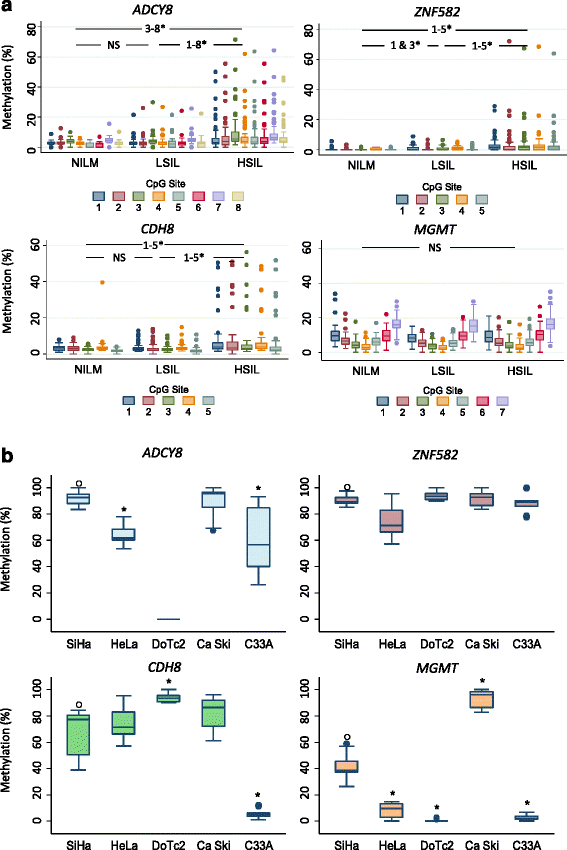
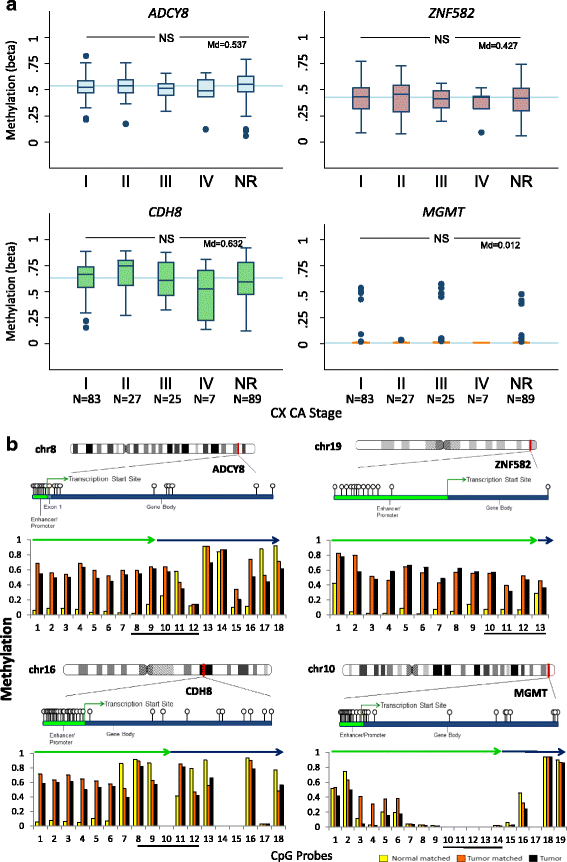
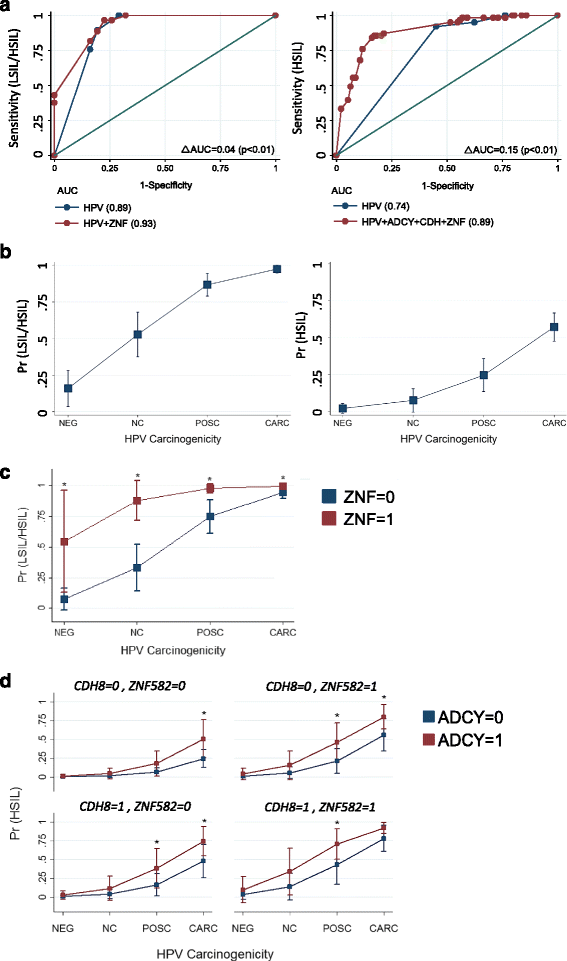
Results: Two hundred seventy-seven quality cytology samples were analyzed. HPV was detected in 31/100 (31 %) negative for intraepithelial lesion or malignancy (NILM), 95/100 (95 %) low-grade squamous intraepithelial lesion (LSIL), and 71/77 (92 %) high-grade squamous intraepithelial lesion (HSIL) samples. The proportion of IARC-defined carcinogenic HPV types in sequenced samples correlated with worsening grade: NILM 7/29 (24 %), LSIL 53/92 (58 %), and HSIL 65/70 (93 %). Promoter methylation of ADCY8, CDH8, and ZNF582 was measured in 170 samples: NILM (N = 33), LSIL (N = 70), and HSIL (N = 67) also correlated with worsening grade. Similar hypermethylation patterns were found in cancer cell lines and TCGA samples. The combination of four biomarkers, i.e., HPV genotype and three-gene promoter methylation, predicted HSIL (AUC 0.89) better than HPV alone (AUC 0.74) by logistic regression and probabilistic modeling.
Conclusions: HPV genotype and DNA methylation of ADCY8, CDH8, and ZNF582 are correlated with cytological grade. Collectively, these biomarkers may serve as a molecular classifier of Pap smears.
Keywords: DNA methylation; HPV; HPV genotyping; Molecular biomarkers; Molecular diagnostics; Pap smear; Pyrosequencing.
Figures
References
-
- Papanicolaou GN, Traut HF. The diagnostic value of vaginal smears in carcinoma of the uterus. Am J Obstet Gynecol. 1941;42:193–206. - PubMed
-
- Carmichael DE, Cameron C. The Pap smear: life of George N. Papanicolaou. Springfield: Charles C. Thomas; 1973. The Pap smear; pp. 68–83.
-
- Organization, World Health . Comprehensive cervical cancer control: a guide to essential practice. 2. Geneva: WHO; 2014. pp. 23–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

