Genome-wide identification and characterization of WRKY gene family in Salix suchowensis
- PMID: 27651997
- PMCID: PMC5018666
- DOI: 10.7717/peerj.2437
Genome-wide identification and characterization of WRKY gene family in Salix suchowensis
Abstract
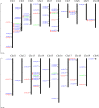
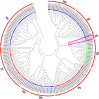
WRKY proteins are the zinc finger transcription factors that were first identified in plants. They can specifically interact with the W-box, which can be found in the promoter region of a large number of plant target genes, to regulate the expressions of downstream target genes. They also participate in diverse physiological and growing processes in plants. Prior to this study, a plenty of WRKY genes have been identified and characterized in herbaceous species, but there is no large-scale study of WRKY genes in willow. With the whole genome sequencing of Salix suchowensis, we have the opportunity to conduct the genome-wide research for willow WRKY gene family. In this study, we identified 85 WRKY genes in the willow genome and renamed them from SsWRKY1 to SsWRKY85 on the basis of their specific distributions on chromosomes. Due to their diverse structural features, the 85 willow WRKY genes could be further classified into three main groups (group I-III), with five subgroups (IIa-IIe) in group II. With the multiple sequence alignment and the manual search, we found three variations of the WRKYGQK heptapeptide: WRKYGRK, WKKYGQK and WRKYGKK, and four variations of the normal zinc finger motif, which might execute some new biological functions. In addition, the SsWRKY genes from the same subgroup share the similar exon-intron structures and conserved motif domains. Further studies of SsWRKY genes revealed that segmental duplication events (SDs) played a more prominent role in the expansion of SsWRKY genes. Distinct expression profiles of SsWRKY genes with RNA sequencing data revealed that diverse expression patterns among five tissues, including tender roots, young leaves, vegetative buds, non-lignified stems and barks. With the analyses of WRKY gene family in willow, it is not only beneficial to complete the functional and annotation information of WRKY genes family in woody plants, but also provide important references to investigate the expansion and evolution of this gene family in flowering plants.
Keywords: Duplication; Evolution; Expression; Phylogenetic analysis; WRKY protein; Willow.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

