Functional versus non-functional intratumor heterogeneity in cancer
- PMID: 27652316
- PMCID: PMC4972105
- DOI: 10.1080/23723556.2016.1162897
Functional versus non-functional intratumor heterogeneity in cancer
Abstract
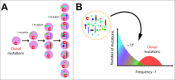
Next-generation sequencing data from human cancers are often difficult to interpret within the context of tumor evolution. We developed a mathematical model describing the accumulation of mutations under neutral evolutionary dynamics and showed that 323/904 cancers (∼30%) from multiple types were consistent with the neutral model of tumor evolution.
Keywords: Cancer genome; intratumor heterogeneity; mathematical model; mutation rate; neutral expansion; site-frequency spectrum.
Figures
References
-
- Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet 2016; 48:3; 238–244; http://dx.doi.org/10.1038/ng.3489 - DOI - PMC - PubMed
-
- Durrett R. Population genetics of neutral mutations in exponentially growing cancer cell populations. Ann Appl Probability 2013; 23:230-50; PMID:23471293; http://dx.doi.org/10.1214/11-AAP824 - DOI - PMC - PubMed
-
- Griffiths RC, Tavaré S. The age of a mutation in a general coalescent tree. Communications in Statistics. Stochastic Models 1998; 14:273-95; http://dx.doi.org/10.1080/15326349808807471 - DOI
-
- Jones S. et al.. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA 2008; 105:4283-8; PMID:18337506; http://dx.doi.org/10.1073/pnas.0712345105 - DOI - PMC - PubMed
-
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990; 61:759-67; PMID:2188735; http://dx.doi.org/10.1016/0092-8674(90)90186-I - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
