Decoding sORF translation - from small proteins to gene regulation
- PMID: 27653973
- PMCID: PMC5100344
- DOI: 10.1080/15476286.2016.1218589
Decoding sORF translation - from small proteins to gene regulation
Abstract
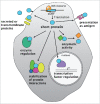
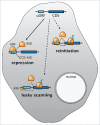
Translation is best known as the fundamental mechanism by which the ribosome converts a sequence of nucleotides into a string of amino acids. Extensive research over many years has elucidated the key principles of translation, and the majority of translated regions were thought to be known. The recent discovery of wide-spread translation outside of annotated protein-coding open reading frames (ORFs) came therefore as a surprise, raising the intriguing possibility that these newly discovered translated regions might have unrecognized protein-coding or gene-regulatory functions. Here, we highlight recent findings that provide evidence that some of these newly discovered translated short ORFs (sORFs) encode functional, previously missed small proteins, while others have regulatory roles. Based on known examples we will also speculate about putative additional roles and the potentially much wider impact that these translated regions might have on cellular homeostasis and gene regulation.
Keywords: Ribosome; sORF; short proteins; translation; translational regulation; uORF.
Figures
References
-
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012; 482:339–46; PMID:22337053; http://dx.doi.org/ 10.1038/nature10887 - DOI - PMC - PubMed
-
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011; 27:i275-82; PMID:21685081; http://dx.doi.org/ 10.1093/bioinformatics/btr209 - DOI - PMC - PubMed
-
- Ingolia NT, Ghaemmaghami S, Newnam JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009; 324:218–23; PMID:19213877; http://dx.doi.org/ 10.1126/science.1168978 - DOI - PMC - PubMed
-
- Ingolia NT, Lareau LF, Weissman JS. Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell 2011; 147(4):1–23; PMID:22056041; http://dx.doi.org/27015305 10.1016/j.cell.2011.10.002 - DOI - PMC - PubMed
-
- Ingolia NT. Ribosome Footprint Profiling of Translation throughout the Genome. Cell 2016; 165:22–33; PMID:27015305; http://dx.doi.org/ 10.1016/j.cell.2016.02.066 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
