tRNA-mimicry in IRES-mediated translation and recoding
- PMID: 27654067
- PMCID: PMC5100355
- DOI: 10.1080/15476286.2016.1219833
tRNA-mimicry in IRES-mediated translation and recoding
Abstract
Viruses maintain compact genomes that must be packaged within capsids typically less than 200 nanometers in diameter. Therefore, instead of coding for a full set of genes needed for replication, viruses have evolved remarkable strategies for co-opting the host cellular machinery. Additionally, viruses often increase the coding capacity of their own genomes by employing overlapping open reading frames (ORFs). Some overlapping viral ORFs involve recoding events that are programmed by the viral RNA. During these programmed recoding events, the ribosome is directed to translate in an alternative reading frame. Here we describe how the Dicistroviridae family of viruses utilize an internal ribosome entry site (IRES) in order to recruit ribosomes to initiate translation at a non-AUG codon. The IRES accomplishes this in part by mimicking the structure of a tRNA. Recently, we showed that the Israeli Acute Paralysis Virus (IAPV) member of the Dicistroviridae family utilizes its IRES to initiate translation in 2 different reading frames. Thus, IAPV has evolved an apparently novel recoding mechanism that reveals important insights into translation. Finally, we compare the IAPV structure to other systems that utilize tRNA mimicry in translation.
Keywords: Frameshifting; IRES; ribosome; translation; virus.
Figures
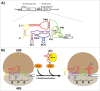
References
-
- Rodnina MV. Quality control of mRNA decoding on the bacterial ribosome. Adv Protein Chem Structural Biol 2012; 86:95-128; PMID:22243582; http://dx.doi.org/ 10.1016/B978-0-12-386497-0.00003-7 - DOI - PubMed
-
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 2004; 13:191-200; PMID:14759365; http://dx.doi.org/ 10.1016/S1097-2765(04)00005-X - DOI - PubMed
-
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 2004; 11:1008-14; PMID:15448679; http://dx.doi.org/ 10.1038/nsmb831 - DOI - PubMed
-
- Dinman JD. Control of gene expression by translational recoding. Adv Protein Chem Structural Biol 2012; 86:129-49; PMID:22243583; http://dx.doi.org/ 10.1016/B978-0-12-386497-0.00004-9 - DOI - PMC - PubMed
-
- Au HH, Cornilescu G, Mouzakis KD, Ren Q, Burke JE, Lee S, Butcher SE, Jan E. Global shape mimicry of tRNA within a viral internal ribosome entry site mediates translational reading frame selection. Proc Natl Acad Sci U S A 2015; 112:E6446-55; PMID:26554019; http://dx.doi.org/ 10.1073/pnas.1512088112 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
