The Cytoskeletal Adaptor Obscurin-Like 1 Interacts with the Human Papillomavirus 16 (HPV16) Capsid Protein L2 and Is Required for HPV16 Endocytosis
- PMID: 27654294
- PMCID: PMC5110159
- DOI: 10.1128/JVI.01222-16
The Cytoskeletal Adaptor Obscurin-Like 1 Interacts with the Human Papillomavirus 16 (HPV16) Capsid Protein L2 and Is Required for HPV16 Endocytosis
Abstract
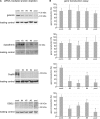
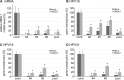
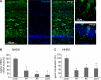
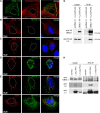
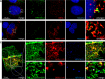
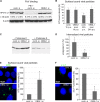
The human papillomavirus (HPV) capsid protein L2 is essential for viral entry. To gain a deeper understanding of the role of L2, we searched for novel cellular L2-interacting proteins. A yeast two-hybrid analysis uncovered the actin-depolymerizing factor gelsolin, the membrane glycoprotein dysadherin, the centrosomal protein 68 (Cep68), and the cytoskeletal adaptor protein obscurin-like 1 protein (OBSL1) as putative L2 binding molecules. Pseudovirus (PsV) infection assays identified OBSL1 as a host factor required for gene transduction by three oncogenic human papillomavirus types, HPV16, HPV18, and HPV31. In addition, we detected OBSL1 expression in cervical tissue sections and noted the involvement of OBSL1 during gene transduction of primary keratinocytes by HPV16 PsV. Complex formation of HPV16 L2 with OBSL1 was demonstrated in coimmunofluorescence and coimmunoprecipitation studies after overexpression of L2 or after PsV exposure. We observed a strong colocalization of OBSL1 with HPV16 PsV and tetraspanin CD151 at the plasma membrane, suggesting a role for OBSL1 in viral endocytosis. Indeed, viral entry assays exhibited a reduction of viral endocytosis in OBSL1-depleted cells. Our results suggest OBSL1 as a novel L2-interacting protein and endocytosis factor in HPV infection.
Importance: Human papillomaviruses infect mucosal and cutaneous epithelia, and the high-risk HPV types account for 5% of cancer cases worldwide. As recently discovered, HPV entry occurs by a clathrin-, caveolin-, and dynamin-independent endocytosis via tetraspanin-enriched microdomains. At present, the cellular proteins involved in the underlying mechanism of this type of endocytosis are under investigation. In this study, the cytoskeletal adaptor OBSL1 was discovered as a previously unrecognized interaction partner of the minor capsid protein L2 and was identified as a proviral host factor required for HPV16 endocytosis into target cells. The findings of this study advance the understanding of a so far less well-characterized endocytic pathway that is used by oncogenic HPV subtypes.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

