The Primary Effect on the Proteome of ARID1A-mutated Ovarian Clear Cell Carcinoma is Downregulation of the Mevalonate Pathway at the Post-transcriptional Level
- PMID: 27654507
- PMCID: PMC5098034
- DOI: 10.1074/mcp.M116.062539
The Primary Effect on the Proteome of ARID1A-mutated Ovarian Clear Cell Carcinoma is Downregulation of the Mevalonate Pathway at the Post-transcriptional Level
Abstract
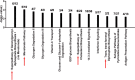
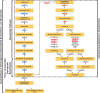
Inactivating mutations in ARID1A, which encodes a subunit of the SWI/SNF chromatin-remodeling complex, are found in over half of ovarian clear cell carcinoma cases and more broadly across most types of cancers. To identify ARID1A-dependent changes in intracellular signaling pathways, we performed proteome analyses of isogenic ovarian clear cell carcinoma cell lines with or without ARID1A expression. Knockout of ARID1A in an ovarian clear cell carcinoma cell line with wild-type ARID1A, OVCA429, primarily resulted in downregulation of the mevalonate pathway, an important metabolic pathway involved in isoprenoid synthesis, cholesterol synthesis, and other downstream pathways. In a complementary experiment, expression of wild-type ARID1A in an ovarian clear cell carcinoma cell line containing mutated ARID1A, OVISE, affected the mevalonate pathway in a reciprocal manner. A striking aspect of these analyses was that, although only 5% of the detected proteome showed significant abundance changes, most proteins in the mevalonate pathway were coordinately affected by ARID1A status. There were generally corresponding changes when comparing the proteomics data to our previously published microarray data for ectopic expression of ARID1A in the OVISE cell line. However, ARID1A-dependent changes were not detected for genes within the mevalonate pathway. This discrepancy suggests that the mevalonate pathway is not regulated directly by ARID1A-mediated transcription and may be regulated post-transcriptionally. We conclude that ARID1A status indirectly influences the mevalonate pathway and probably influences other processes including glycogen metabolism and 14-3-3-mediated signaling. Further, our findings demonstrate that changes in mRNA levels are sometimes poor indicators of signaling pathways affected by gene manipulations in cancer cells.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
ARID1A mutations in endometriosis-associated ovarian carcinomas.N Engl J Med. 2010 Oct 14;363(16):1532-43. doi: 10.1056/NEJMoa1008433. Epub 2010 Sep 8. N Engl J Med. 2010. PMID: 20942669 Free PMC article.
-
Targeting the mevalonate pathway suppresses ARID1A-inactivated cancers by promoting pyroptosis.Cancer Cell. 2023 Apr 10;41(4):740-756.e10. doi: 10.1016/j.ccell.2023.03.002. Epub 2023 Mar 23. Cancer Cell. 2023. PMID: 36963401 Free PMC article.
-
A functional proteogenomic analysis of endometrioid and clear cell carcinomas using reverse phase protein array and mutation analysis: protein expression is histotype-specific and loss of ARID1A/BAF250a is associated with AKT phosphorylation.BMC Cancer. 2014 Feb 22;14:120. doi: 10.1186/1471-2407-14-120. BMC Cancer. 2014. PMID: 24559118 Free PMC article.
-
Potential therapeutic targets in ARID1A-mutated cancers.Expert Opin Ther Targets. 2015;19(11):1419-22. doi: 10.1517/14728222.2015.1062879. Epub 2015 Jun 30. Expert Opin Ther Targets. 2015. PMID: 26125128 Free PMC article. Review.
-
ARID1A mutant ovarian clear cell carcinoma: A clear target for synthetic lethal strategies.Biochim Biophys Acta Rev Cancer. 2018 Dec;1870(2):176-184. doi: 10.1016/j.bbcan.2018.07.005. Epub 2018 Jul 17. Biochim Biophys Acta Rev Cancer. 2018. PMID: 30025943 Review.
Cited by
-
Changes in the urinary proteome before and after quadrivalent influenza vaccine and COVID-19 vaccination.Front Immunol. 2022 Oct 7;13:946791. doi: 10.3389/fimmu.2022.946791. eCollection 2022. Front Immunol. 2022. PMID: 36275736 Free PMC article.
-
Treatment for ovarian clear cell carcinoma with combined inhibition of WEE1 and ATR.J Ovarian Res. 2023 Apr 22;16(1):80. doi: 10.1186/s13048-023-01160-y. J Ovarian Res. 2023. PMID: 37087441 Free PMC article.
-
The Mevalonate Pathway, a Metabolic Target in Cancer Therapy.Front Oncol. 2021 Feb 25;11:626971. doi: 10.3389/fonc.2021.626971. eCollection 2021. Front Oncol. 2021. PMID: 33718197 Free PMC article. Review.
-
A High-Density Genetic Linkage Map and Fine Mapping of QTL For Feed Conversion Efficiency in Common Carp (Cyprinus carpio).Front Genet. 2021 Nov 12;12:778487. doi: 10.3389/fgene.2021.778487. eCollection 2021. Front Genet. 2021. PMID: 34868267 Free PMC article.
-
The Tumor Suppressor ARID1A Controls Global Transcription via Pausing of RNA Polymerase II.Cell Rep. 2018 Jun 26;23(13):3933-3945. doi: 10.1016/j.celrep.2018.05.097. Cell Rep. 2018. PMID: 29949775 Free PMC article.
References
-
- Siegel R. L., Miller K. D., and Jemal A. (2016) Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30 - PubMed
-
- Pectasides D., Pectasides E., Psyrri A., and Economopoulos T. (2006) Treatment issues in clear cell carcinoma of the ovary: a different entity? Oncologist 11, 1089–1094 - PubMed
-
- Yamaguchi K., Mandai M., Oura T., Matsumura N., Hamanishi J., Baba T., Matsui S., Murphy S. K., and Konishi I. (2010) Identification of an ovarian clear cell carcinoma gene signature that reflects inherent disease biology and the carcinogenic processes. Oncogene 29, 1741–1752 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

