Inhibition of soluble guanylyl cyclase by small molecules targeting the catalytic domain
- PMID: 27654641
- PMCID: PMC5077689
- DOI: 10.1002/1873-3468.12427
Inhibition of soluble guanylyl cyclase by small molecules targeting the catalytic domain
Abstract
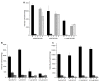
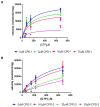
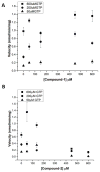
Soluble guanylyl cyclase (sGC) plays a crucial role in cyclic nucleotide signaling that regulates numerous important physiological processes. To identify new sGC inhibitors that may prevent the formation of the active catalytic domain conformation, we carried out an in silico docking screen targeting a 'backside pocket' of the inactive sGC catalytic domain structure. Compounds 1 and 2 were discovered to inhibit sGC even at high/saturating nitric oxide concentrations. Both compounds also inhibit the BAY 58-2667-activated sGC as well as BAY 41-2272-stimulated sGC activity. Additional biochemical analyses showed that compound 2 also inhibits the isolated catalytic domain, thus demonstrating functional binding to this domain. Both compounds have micromolar affinity for sGC and are potential leads to develop more potent sGC inhibitors.
Keywords: enzyme inhibition; soluble guanylyl cyclase.
© 2016 Federation of European Biochemical Societies.
Figures
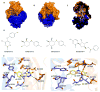


References
-
- Katsuki S, Arnold W, Mittal C, Murad F. Stimulation of guanylate cyclase by sodium nitroprusside, nitroglycerin and nitric oxide in various tissue preparations and comparison to the effects of sodium azide and hydroxylamine. J Cyclic Nucleotide Res. 1977;3:23–35. - PubMed
-
- Denninger JW, Marletta MA. Guanylate cyclase and the .NO/cGMP signaling pathway. Biochim Biophys Acta. 1999;1411:334–350. - PubMed
-
- Humbert P, Niroomand F, Fischer G, Mayer B, Koesling D, Hinsch KD, Gausepohl H, Frank R, Schultz G, Bohme E. Purification of soluble guanylyl cyclase from bovine lung by a new immunoaffinity chromatographic method. Eur J Biochem. 1990;190:273–278. - PubMed
-
- Stone JR, Marletta MA. Spectral and kinetic studies on the activation of soluble guanylate cyclase by nitric oxide. Biochemistry. 1996;35:1093–1099. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

