Cancer biomarker discovery is improved by accounting for variability in general levels of drug sensitivity in pre-clinical models
- PMID: 27654937
- PMCID: PMC5031330
- DOI: 10.1186/s13059-016-1050-9
Cancer biomarker discovery is improved by accounting for variability in general levels of drug sensitivity in pre-clinical models
Abstract
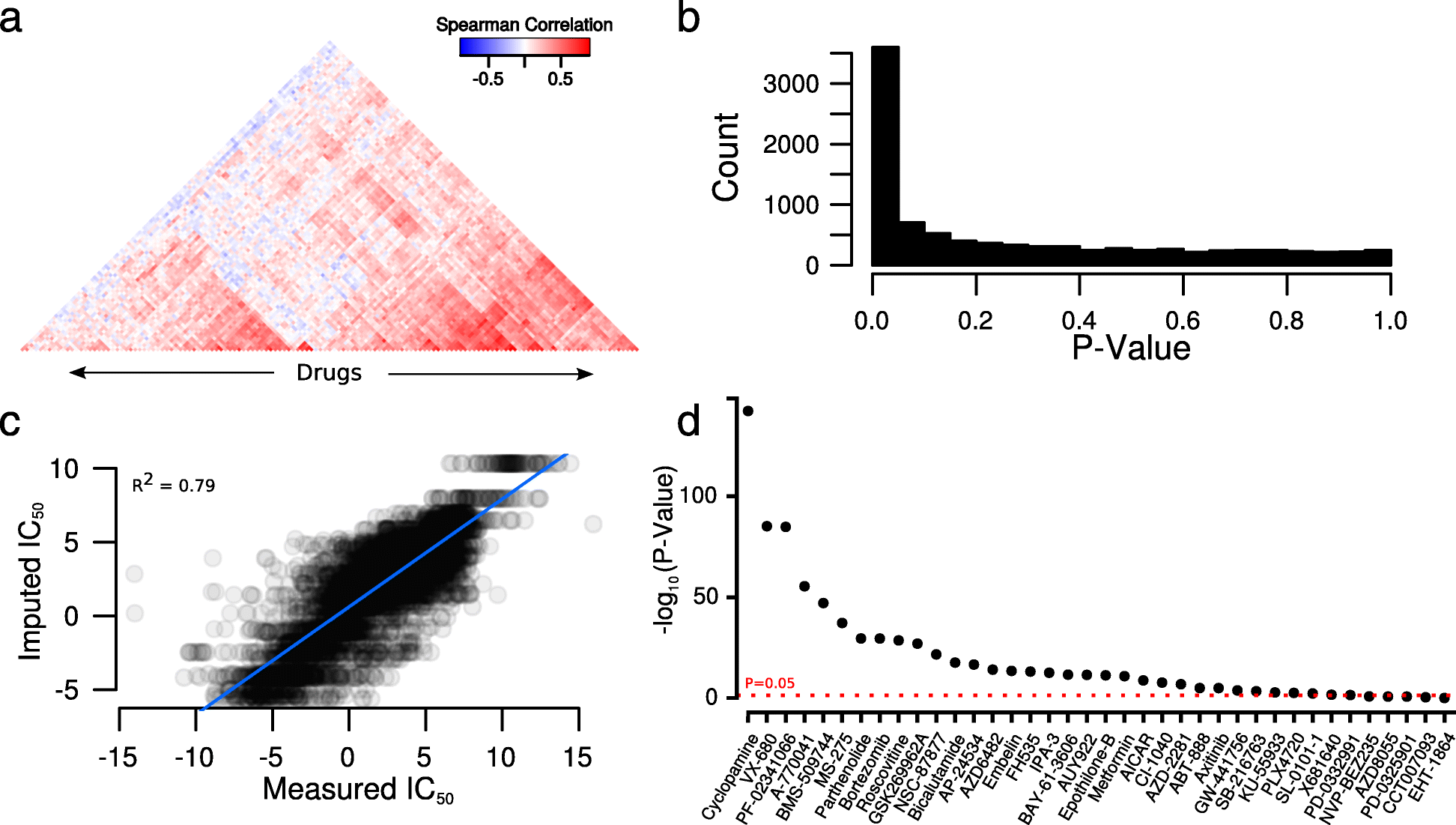
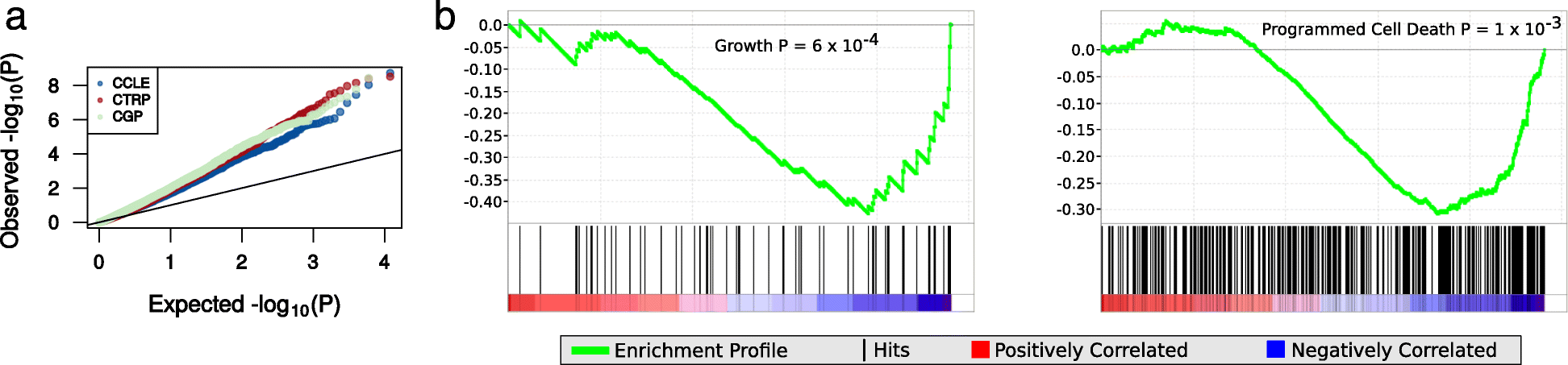
We show that variability in general levels of drug sensitivity in pre-clinical cancer models confounds biomarker discovery. However, using a very large panel of cell lines, each treated with many drugs, we could estimate a general level of sensitivity to all drugs in each cell line. By conditioning on this variable, biomarkers were identified that were more likely to be effective in clinical trials than those identified using a conventional uncorrected approach. We find that differences in general levels of drug sensitivity are driven by biologically relevant processes. We developed a gene expression based method that can be used to correct for this confounder in future studies.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

