Serum depletion induced cancer stem cell-like phenotype due to nitric oxide synthesis in oncogenic HRas transformed cells
- PMID: 27655692
- PMCID: PMC5342736
- DOI: 10.18632/oncotarget.12117
Serum depletion induced cancer stem cell-like phenotype due to nitric oxide synthesis in oncogenic HRas transformed cells
Abstract
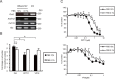
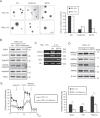
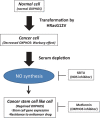
Cancer cells rewire their metabolism and mitochondrial oxidative phosphorylation (OXPHOS) to promote proliferation and maintenance. Cancer cells use multiple adaptive mechanisms in response to a hypo-nutrient environment. However, little is known about how cancer mitochondria are involved in the ability of these cells to adapt to a hypo-nutrient environment. Oncogenic HRas leads to suppression of the mitochondrial oxygen consumption rate (OCR), but oxygen consumption is essential for tumorigenesis. We found that in oncogenic HRas transformed cells, serum depletion reversibly increased the OCR and membrane potential. Serum depletion promoted a cancer stem cell (CSC)-like phenotype, indicated by an increase in CSC markers expression and resistance to anticancer agents. We also found that nitric oxide (NO) synthesis was significantly induced after serum depletion and that NO donors modified the OCR. An NOS inhibitor, SEITU, inhibited the OCR and CSC gene expression. It also reduced anchorage-independent growth by promoting apoptosis. In summary, our data provide new molecular findings that serum depletion induces NO synthesis and promotes mitochondrial OXPHOS, leading to tumor progression and a CSC phenotype. These results suggest that mitochondrial OCR inhibitors can be used as therapy against CSC.
Keywords: HRas; OXPHOS; cancer stem cell; nitric oxide; serum depletion.
Conflict of interest statement
The authors declare there are no potential conflicts of interest to disclose.
Figures




References
-
- Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol. 2005;70:363–374. - PubMed
-
- DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. - PubMed
-
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. - PubMed
-
- Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

