How curvature-generating proteins build scaffolds on membrane nanotubes
- PMID: 27655892
- PMCID: PMC5056078
- DOI: 10.1073/pnas.1606943113
How curvature-generating proteins build scaffolds on membrane nanotubes
Abstract
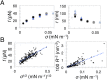
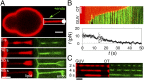
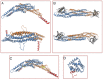
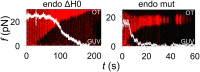
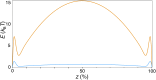
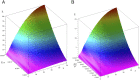
Bin/Amphiphysin/Rvs (BAR) domain proteins control the curvature of lipid membranes in endocytosis, trafficking, cell motility, the formation of complex subcellular structures, and many other cellular phenomena. They form 3D assemblies that act as molecular scaffolds to reshape the membrane and alter its mechanical properties. It is unknown, however, how a protein scaffold forms and how BAR domains interact in these assemblies at protein densities relevant for a cell. In this work, we use various experimental, theoretical, and simulation approaches to explore how BAR proteins organize to form a scaffold on a membrane nanotube. By combining quantitative microscopy with analytical modeling, we demonstrate that a highly curving BAR protein endophilin nucleates its scaffolds at the ends of a membrane tube, contrary to a weaker curving protein centaurin, which binds evenly along the tube's length. Our work implies that the nature of local protein-membrane interactions can affect the specific localization of proteins on membrane-remodeling sites. Furthermore, we show that amphipathic helices are dispensable in forming protein scaffolds. Finally, we explore a possible molecular structure of a BAR-domain scaffold using coarse-grained molecular dynamics simulations. Together with fluorescence microscopy, the simulations show that proteins need only to cover 30-40% of a tube's surface to form a rigid assembly. Our work provides mechanical and structural insights into the way BAR proteins may sculpt the membrane as a high-order cooperative assembly in important biological processes.
Keywords: BAR proteins; endocytosis; membrane curvature; protein scaffold; self-assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





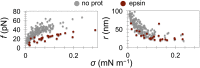


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

