A Review of SHV Extended-Spectrum β-Lactamases: Neglected Yet Ubiquitous
- PMID: 27656166
- PMCID: PMC5011133
- DOI: 10.3389/fmicb.2016.01374
A Review of SHV Extended-Spectrum β-Lactamases: Neglected Yet Ubiquitous
Abstract
β-lactamases are the primary cause of resistance to β-lactams among members of the family Enterobacteriaceae. SHV enzymes have emerged in Enterobacteriaceae causing infections in health care in the last decades of the Twentieth century, and they are now observed in isolates in different epidemiological settings both in human, animal and the environment. Likely originated from a chromosomal penicillinase of Klebsiella pneumoniae, SHV β-lactamases currently encompass a large number of allelic variants including extended-spectrum β-lactamases (ESBL), non-ESBL and several not classified variants. SHV enzymes have evolved from a narrow- to an extended-spectrum of hydrolyzing activity, including monobactams and carbapenems, as a result of amino acid changes that altered the configuration around the active site of the β -lactamases. SHV-ESBLs are usually encoded by self-transmissible plasmids that frequently carry resistance genes to other drug classes and have become widespread throughout the world in several Enterobacteriaceae, emphasizing their clinical significance.
Keywords: ESBL; Enterobacteriaceae; SHV-12; SHV-2; SHV-5; blaSHV; plasmid; β-lactamase.
Figures
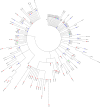
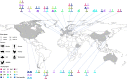

References
-
- Abbassi M. S., Torres C., Achour W., Vinué L., Sáenz Y., Costa D., et al. (2008). Genetic characterisation of CTX-M-15-producing Klebsiella pneumoniae and Escherichia coli strains isolated from stem cell transplant patients in Tunisia. Int. J. Antimicrob. Agents 32, 308–314. 10.1016/j.ijantimicag.2008.04.009 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

