Genome Analysis and Characterisation of the Exopolysaccharide Produced by Bifidobacterium longum subsp. longum 35624™
- PMID: 27656878
- PMCID: PMC5033381
- DOI: 10.1371/journal.pone.0162983
Genome Analysis and Characterisation of the Exopolysaccharide Produced by Bifidobacterium longum subsp. longum 35624™
Abstract
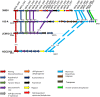
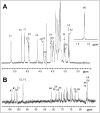
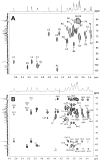
The Bifibobacterium longum subsp. longum 35624™ strain (formerly named Bifidobacterium longum subsp. infantis) is a well described probiotic with clinical efficacy in Irritable Bowel Syndrome clinical trials and induces immunoregulatory effects in mice and in humans. This paper presents (a) the genome sequence of the organism allowing the assignment to its correct subspeciation longum; (b) a comparative genome assessment with other B. longum strains and (c) the molecular structure of the 35624 exopolysaccharide (EPS624). Comparative genome analysis of the 35624 strain with other B. longum strains determined that the sub-speciation of the strain is longum and revealed the presence of a 35624-specific gene cluster, predicted to encode the biosynthetic machinery for EPS624. Following isolation and acid treatment of the EPS, its chemical structure was determined using gas and liquid chromatography for sugar constituent and linkage analysis, electrospray and matrix assisted laser desorption ionization mass spectrometry for sequencing and NMR. The EPS consists of a branched hexasaccharide repeating unit containing two galactose and two glucose moieties, galacturonic acid and the unusual sugar 6-deoxy-L-talose. These data demonstrate that the B. longum 35624 strain has specific genetic features, one of which leads to the generation of a characteristic exopolysaccharide.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: David Groeger, Elisa Schiavi and Ray Grant are employees of Alimentary Health Pharma Davos AG, Jun Xu is an employee of Procter & Gamble, while Stephan Plattner, Selena Healy and Jennifer Roper are employees of Alimentary Health Ltd. Liam O’Mahony has received research funding from GSK and is a consultant to Alimentary Health Ltd. Cezmi Akdis has received research support from Novartis and Stallergenes and consulted for Actellion, Aventis and Allergopharma. Friedrich Altmann, Paul Kosma, Elisabeth Svehla, Markus Windwarder, Andreas Hofinger, Amy O’Callaghan, Sinead Leahy, Francesca Bottacini, Evelyn Molloy, Noelia Rodriguez Perez, Elisa Schiavi, Marita Gleinser, Mary O’Connell Motherway and Douwe van Sinderen have no competing interests. While competing interests are declared for some of the authors, the content of this article was neither influenced nor constrained by this fact and this does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures





References
-
- Frei R, Akdis M, O'Mahony L. Prebiotics, probiotics, synbiotics, and the immune system: experimental data and clinical evidence. Curr Opin Gastroenterol 2015;31: 153–158. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

