Functional Equivalence of Retroviral MA Domains in Facilitating Psi RNA Binding Specificity by Gag
- PMID: 27657107
- PMCID: PMC5035970
- DOI: 10.3390/v8090256
Functional Equivalence of Retroviral MA Domains in Facilitating Psi RNA Binding Specificity by Gag
Abstract
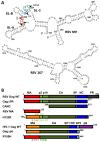
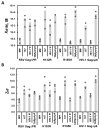
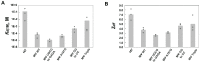
Retroviruses specifically package full-length, dimeric genomic RNA (gRNA) even in the presence of a vast excess of cellular RNA. The "psi" (Ψ) element within the 5'-untranslated region (5'UTR) of gRNA is critical for packaging through interaction with the nucleocapsid (NC) domain of Gag. However, in vitro Gag binding affinity for Ψ versus non-Ψ RNAs is not significantly different. Previous salt-titration binding assays revealed that human immunodeficiency virus type 1 (HIV-1) Gag bound to Ψ RNA with high specificity and relatively few charge interactions, whereas binding to non-Ψ RNA was less specific and involved more electrostatic interactions. The NC domain was critical for specific Ψ binding, but surprisingly, a Gag mutant lacking the matrix (MA) domain was less effective at discriminating Ψ from non-Ψ RNA. We now find that Rous sarcoma virus (RSV) Gag also effectively discriminates RSV Ψ from non-Ψ RNA in a MA-dependent manner. Interestingly, Gag chimeras, wherein the HIV-1 and RSV MA domains were swapped, maintained high binding specificity to cognate Ψ RNAs. Using Ψ RNA mutant constructs, determinants responsible for promoting high Gag binding specificity were identified in both systems. Taken together, these studies reveal the functional equivalence of HIV-1 and RSV MA domains in facilitating Ψ RNA selectivity by Gag, as well as Ψ elements that promote this selectivity.
Keywords: Gag; Psi RNA (Ψ RNA); Rous sarcoma virus (RSV); human immunodeficiency virus type 1 (HIV-1); matrix (MA); nucleocapsid (NC).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging.RNA. 2013 Aug;19(8):1078-88. doi: 10.1261/rna.038869.113. Epub 2013 Jun 24. RNA. 2013. PMID: 23798665 Free PMC article.
-
Mechanistic differences between nucleic acid chaperone activities of the Gag proteins of Rous sarcoma virus and human immunodeficiency virus type 1 are attributed to the MA domain.J Virol. 2014 Jul;88(14):7852-61. doi: 10.1128/JVI.00736-14. Epub 2014 Apr 30. J Virol. 2014. PMID: 24789780 Free PMC article.
-
The matrix domain contributes to the nucleic acid chaperone activity of HIV-2 Gag.Retrovirology. 2016 Mar 17;13:18. doi: 10.1186/s12977-016-0245-1. Retrovirology. 2016. PMID: 26987314 Free PMC article.
-
Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly.Semin Cell Dev Biol. 2019 Feb;86:129-139. doi: 10.1016/j.semcdb.2018.03.015. Epub 2018 Apr 1. Semin Cell Dev Biol. 2019. PMID: 29580971 Free PMC article. Review.
-
On the Selective Packaging of Genomic RNA by HIV-1.Viruses. 2016 Sep 12;8(9):246. doi: 10.3390/v8090246. Viruses. 2016. PMID: 27626441 Free PMC article. Review.
Cited by
-
The Rous sarcoma virus Gag Polyprotein Forms Biomolecular Condensates Driven by Intrinsically-disordered Regions.J Mol Biol. 2023 Aug 15;435(16):168182. doi: 10.1016/j.jmb.2023.168182. Epub 2023 Jun 14. J Mol Biol. 2023. PMID: 37328094 Free PMC article.
-
HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites.Viruses. 2020 Nov 9;12(11):1281. doi: 10.3390/v12111281. Viruses. 2020. PMID: 33182496 Free PMC article.
-
Mechanistic differences between HIV-1 and SIV nucleocapsid proteins and cross-species HIV-1 genomic RNA recognition.Retrovirology. 2016 Dec 29;13(1):89. doi: 10.1186/s12977-016-0322-5. Retrovirology. 2016. PMID: 28034301 Free PMC article.
-
Mutations in the Basic Region of the Mason-Pfizer Monkey Virus Nucleocapsid Protein Affect Reverse Transcription, Genomic RNA Packaging, and the Virus Assembly Site.J Virol. 2018 Apr 27;92(10):e00106-18. doi: 10.1128/JVI.00106-18. Print 2018 May 15. J Virol. 2018. PMID: 29491167 Free PMC article.
-
Transcription start site choice regulates HIV-1 RNA conformation and function.Curr Opin Struct Biol. 2024 Oct;88:102896. doi: 10.1016/j.sbi.2024.102896. Epub 2024 Aug 14. Curr Opin Struct Biol. 2024. PMID: 39146887 Review.
References
-
- Berkowitz R., Fisher J., Goff S.P. RNA packaging. Curr. Top. Microbiol. Immunol. 1996;214:177–218. - PubMed
-
- Chen J., Nikolaitchik O., Singh J., Wright A., Bencsics C.E., Coffin J.M., Ni N., Lockett S., Pathak V.K., Hu W.S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. USA. 2009;106:13535–13540. doi: 10.1073/pnas.0906822106. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

