Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors
- PMID: 27657110
- PMCID: PMC5035971
- DOI: 10.3390/v8090257
Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors
Abstract
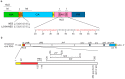
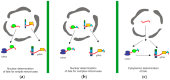
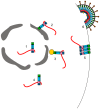
Infectious retrovirus particles contain two copies of unspliced viral RNA that serve as the viral genome. Unspliced retroviral RNA is transcribed in the nucleus by the host RNA polymerase II and has three potential fates: (1) it can be spliced into subgenomic messenger RNAs (mRNAs) for the translation of viral proteins; or it can remain unspliced to serve as either (2) the mRNA for the translation of Gag and Gag-Pol; or (3) the genomic RNA (gRNA) that is packaged into virions. The Gag structural protein recognizes and binds the unspliced viral RNA to select it as a genome, which is selected in preference to spliced viral RNAs and cellular RNAs. In this review, we summarize the current state of understanding about how retroviral packaging is orchestrated within the cell and explore potential new mechanisms based on recent discoveries in the field. We discuss the cis-acting elements in the unspliced viral RNA and the properties of the Gag protein that are required for their interaction. In addition, we discuss the role of host factors in influencing the fate of the newly transcribed viral RNA, current models for how retroviruses distinguish unspliced viral mRNA from viral genomic RNA, and the possible subcellular sites of genomic RNA dimerization and selection by Gag. Although this review centers primarily on the wealth of data available for the alpharetrovirus Rous sarcoma virus, in which a discrete RNA packaging sequence has been identified, we have also summarized the cis- and trans-acting factors as well as the mechanisms governing gRNA packaging of other retroviruses for comparison.
Keywords: retroviral Gag proteins; retrovirus assembly; retroviruses; subcellular trafficking; viral RNA export; viral RNA packaging; virus–cell interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Stoltzfus C.M. Synthesis and processing of avian sarcoma retrovirus RNA. Adv. Virus Res. 1988;35:1–38. - PubMed
-
- Coffin J.M., Hughes S.H., Varmus H.E. The interactions of retroviruses and their hosts. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Cold Spring Harbor; New York, NY, USA: 1997. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

