NDST1 Preferred Promoter Confirmation and Identification of Corresponding Transcriptional Inhibitors as Substrate Reduction Agents for Multiple Mucopolysaccharidosis Disorders
- PMID: 27657498
- PMCID: PMC5033324
- DOI: 10.1371/journal.pone.0162145
NDST1 Preferred Promoter Confirmation and Identification of Corresponding Transcriptional Inhibitors as Substrate Reduction Agents for Multiple Mucopolysaccharidosis Disorders
Abstract
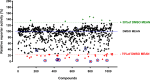
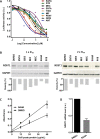
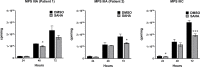
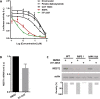
The stepwise degradation of glycosaminoglycans (GAGs) is accomplished by twelve lysosomal enzymes. Deficiency in any of these enzymes will result in the accumulation of the intermediate substrates on the pathway to the complete turnover of GAGs. The accumulation of these undegraded substrates in almost any tissue is a hallmark of all Mucopolysaccharidoses (MPS). Present therapeutics based on enzyme replacement therapy and bone marrow transplantation have low effectiveness for the treatment of MPS with neurological complications since enzymes used in these therapies are unable to cross the blood brain barrier. Small molecule-based approaches are more promising in addressing neurological manifestations. In this report we identify a target for developing a substrate reduction therapy (SRT) for six MPS resulting from the abnormal degradation of heparan sulfate (HS). Using the minimal promoter of NDST1, one of the first modifying enzymes of HS precursors, we established a luciferase based reporter gene assay capable of identifying small molecules that could potentially reduce HS maturation and therefore lessen HS accumulation in certain MPS. From the screen of 1,200 compounds comprising the Prestwick Chemical library we identified SAHA, a histone deacetylase inhibitor, as the drug that produced the highest inhibitory effects in the reporter assay. More importantly SAHA treated fibroblasts expressed lower levels of endogenous NDST1 and accumulated less 35S GAGs in patient cells. Thus, by using our simple reporter gene assay we have demonstrated that by inhibiting the transcription of NDST1 with small molecules, identified by high throughput screening, we can also reduce the level of sulfated HS substrate in MPS patient cells, potentially leading to SRT.
Conflict of interest statement
The authors have declared that no competing interest exist.
Figures


Similar articles
-
Putative biological mechanisms of efficiency of substrate reduction therapies for mucopolysaccharidoses.Arch Immunol Ther Exp (Warsz). 2012 Dec;60(6):461-8. doi: 10.1007/s00005-012-0195-9. Epub 2012 Sep 5. Arch Immunol Ther Exp (Warsz). 2012. PMID: 22949095 Review.
-
Targeting Heparan Sulfate Proteoglycans as a Novel Therapeutic Strategy for Mucopolysaccharidoses.Mol Ther Methods Clin Dev. 2018 Jun 18;10:8-16. doi: 10.1016/j.omtm.2018.05.002. eCollection 2018 Sep 21. Mol Ther Methods Clin Dev. 2018. PMID: 29942826 Free PMC article.
-
A straightforward, quantitative ultra-performance liquid chromatography-tandem mass spectrometric method for heparan sulfate, dermatan sulfate and chondroitin sulfate in urine: an improved clinical screening test for the mucopolysaccharidoses.Mol Genet Metab. 2015 Feb;114(2):123-8. doi: 10.1016/j.ymgme.2014.09.009. Epub 2014 Oct 5. Mol Genet Metab. 2015. PMID: 25458519
-
Separation of sulfated urinary glycosaminoglycans by high-resolution electrophoresis for isotyping of mucopolysaccharidoses in Malaysia.Malays J Pathol. 2010 Jun;32(1):35-42. Malays J Pathol. 2010. PMID: 20614724
-
Mucopolysaccharidoses and the eye.Surv Ophthalmol. 2006 Jan-Feb;51(1):1-17. doi: 10.1016/j.survophthal.2005.11.007. Surv Ophthalmol. 2006. PMID: 16414358 Review.
Cited by
-
Plant chemical genetics reveals colistin sulphate as a SA and NPR1-independent PR1 inducer functioning via a p38-like kinase pathway.Sci Rep. 2019 Aug 1;9(1):11196. doi: 10.1038/s41598-019-47526-5. Sci Rep. 2019. PMID: 31371749 Free PMC article.
-
Glycocalyx mechanotransduction mechanisms are involved in renal cancer metastasis.Matrix Biol Plus. 2022 Jan 6;13:100100. doi: 10.1016/j.mbplus.2021.100100. eCollection 2022 Feb. Matrix Biol Plus. 2022. PMID: 35106474 Free PMC article.
-
Substrate reduction therapy for inborn errors of metabolism.Emerg Top Life Sci. 2019 Mar 29;3(1):63-73. doi: 10.1042/ETLS20180058. Emerg Top Life Sci. 2019. PMID: 33523197 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

