An MSC2 Promoter-lacZ Fusion Gene Reveals Zinc-Responsive Changes in Sites of Transcription Initiation That Occur across the Yeast Genome
- PMID: 27657924
- PMCID: PMC5033525
- DOI: 10.1371/journal.pone.0163256
An MSC2 Promoter-lacZ Fusion Gene Reveals Zinc-Responsive Changes in Sites of Transcription Initiation That Occur across the Yeast Genome
Abstract
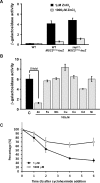
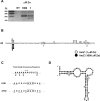
The Msc2 and Zrg17 proteins of Saccharomyces cerevisiae form a complex to transport zinc into the endoplasmic reticulum. ZRG17 is transcriptionally induced in zinc-limited cells by the Zap1 transcription factor. In this report, we show that MSC2 mRNA also increases (~1.5 fold) in zinc-limited cells. The MSC2 gene has two in-frame ATG codons at its 5' end, ATG1 and ATG2; ATG2 is the predicted initiation codon. When the MSC2 promoter was fused at ATG2 to the lacZ gene, we found that unlike the chromosomal gene this reporter showed a 4-fold decrease in lacZ mRNA in zinc-limited cells. Surprisingly, β-galactosidase activity generated by this fusion gene increased ~7 fold during zinc deficiency suggesting the influence of post-transcriptional factors. Transcription of MSC2ATG2-lacZ was found to start upstream of ATG1 in zinc-replete cells. In zinc-limited cells, transcription initiation shifted to sites just upstream of ATG2. From the results of mutational and polysome profile analyses, we propose the following explanation for these effects. In zinc-replete cells, MSC2ATG2-lacZ mRNA with long 5' UTRs fold into secondary structures that inhibit translation. In zinc-limited cells, transcripts with shorter unstructured 5' UTRs are generated that are more efficiently translated. Surprisingly, chromosomal MSC2 did not show start site shifts in response to zinc status and only shorter 5' UTRs were observed. However, the shifts that occur in the MSC2ATG2-lacZ construct led us to identify significant transcription start site changes affecting the expression of ~3% of all genes. Therefore, zinc status can profoundly alter transcription initiation across the yeast genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



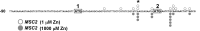


Similar articles
-
The Zap1 transcriptional activator negatively regulates translation of the RTC4 mRNA through the use of alternative 5' transcript leaders.Mol Microbiol. 2017 Dec;106(5):673-677. doi: 10.1111/mmi.13856. Epub 2017 Oct 26. Mol Microbiol. 2017. PMID: 28971534 Free PMC article.
-
Transcriptional regulation of the Zrg17 zinc transporter of the yeast secretory pathway.Biochem J. 2011 Apr 1;435(1):259-66. doi: 10.1042/BJ20102003. Biochem J. 2011. PMID: 21250939 Free PMC article.
-
Zap1-dependent transcription from an alternative upstream promoter controls translation of RTC4 mRNA in zinc-deficient Saccharomyces cerevisiae.Mol Microbiol. 2017 Dec;106(5):678-689. doi: 10.1111/mmi.13851. Epub 2017 Oct 23. Mol Microbiol. 2017. PMID: 28963784 Free PMC article.
-
Changes in transcription start sites of Zap1-regulated genes during zinc deficiency: Implications for HNT1 gene regulation.Mol Microbiol. 2020 Jan;113(1):285-296. doi: 10.1111/mmi.14416. Epub 2019 Nov 24. Mol Microbiol. 2020. PMID: 31692084 Free PMC article.
-
Expression of the Saccharomyces cerevisiae PIS1 gene is modulated by multiple ATGs in the promoter.Biochem Biophys Res Commun. 2006 Nov 10;350(1):91-6. doi: 10.1016/j.bbrc.2006.08.196. Epub 2006 Sep 15. Biochem Biophys Res Commun. 2006. PMID: 16997274
Cited by
-
Structures, Mechanisms, and Physiological Functions of Zinc Transporters in Different Biological Kingdoms.Int J Mol Sci. 2024 Mar 6;25(5):3045. doi: 10.3390/ijms25053045. Int J Mol Sci. 2024. PMID: 38474291 Free PMC article. Review.
-
The GIS2 Gene Is Repressed by a Zinc-Regulated Bicistronic RNA in Saccharomyces cerevisiae.Genes (Basel). 2018 Sep 19;9(9):462. doi: 10.3390/genes9090462. Genes (Basel). 2018. PMID: 30235899 Free PMC article.
-
The Zap1 transcriptional activator negatively regulates translation of the RTC4 mRNA through the use of alternative 5' transcript leaders.Mol Microbiol. 2017 Dec;106(5):673-677. doi: 10.1111/mmi.13856. Epub 2017 Oct 26. Mol Microbiol. 2017. PMID: 28971534 Free PMC article.
-
The interactome of the Bakers' yeast peroxiredoxin Tsa1 implicates it in the redox regulation of intermediary metabolism, glycolysis and zinc homeostasis.bioRxiv [Preprint]. 2025 Feb 21:2025.02.18.638137. doi: 10.1101/2025.02.18.638137. bioRxiv. 2025. Update in: Metallomics. 2025 Aug 5;17(8):mfaf028. doi: 10.1093/mtomcs/mfaf028. PMID: 40027620 Free PMC article. Updated. Preprint.
-
Zinc at the Host-Fungus Interface: How to Uptake the Metal?J Fungi (Basel). 2020 Nov 21;6(4):305. doi: 10.3390/jof6040305. J Fungi (Basel). 2020. PMID: 33233335 Free PMC article. Review.
References
-
- Ellis CD, Macdiarmid CW, Eide DJ (2005) Heteromeric protein complexes mediate zinc transport into the secretory pathway of eukaryotic cells. J Biol Chem 280: 28811–28818. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

