The High-Risk Human Papillomavirus E6 Oncogene Exacerbates the Negative Effect of Tryptophan Starvation on the Development of Chlamydia trachomatis
- PMID: 27658027
- PMCID: PMC5033384
- DOI: 10.1371/journal.pone.0163174
The High-Risk Human Papillomavirus E6 Oncogene Exacerbates the Negative Effect of Tryptophan Starvation on the Development of Chlamydia trachomatis
Abstract
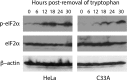
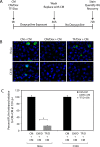
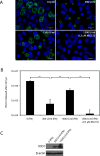
Chlamydia trachomatis is an obligate intracellular pathogen that requires specific essential nutrients from the host cell, one of which is the amino acid tryptophan. In this context interferon gamma (IFNγ) is the major host protective cytokine against chlamydial infections because it induces the expression of the host enzyme, indoleamine 2,3-dioxygenase 1, that degrades tryptophan, thereby restricting bacterial replication. The mechanism by which IFNγ acts has been dissected in vitro using epithelial cell-lines such as HeLa, HEp-2, or the primary-like endocervical cell-line A2EN. All these cell-lines express the high-risk human papillomavirus oncogenes E6 & E7. While screening cell-lines to identify those suitable for C. trachomatis co-infections with other genital pathogens, we unexpectedly found that tryptophan starvation did not completely block chlamydial development in cell-lines that were HR-HPV negative, such as C33A and 293. Therefore, we tested the hypothesis that HR-HPV oncogenes modulate the effect of tryptophan starvation on chlamydial development by comparing chlamydial development in HeLa and C33A cell-lines that were both derived from cervical carcinomas. Our results indicate that during tryptophan depletion, unlike HeLa, C33A cells generate sufficient intracellular tryptophan via proteasomal activity to permit C. trachomatis replication. By generating stable derivatives of C33A that expressed HPV16 E6, E7 or E6 & E7, we found that E6 expression alone was sufficient to convert C33A cells to behave like HeLa during tryptophan starvation. The reduced tryptophan levels in HeLa cells have a biological consequence; akin to the previously described effect of IFNγ, tryptophan starvation protects C. trachomatis from clearance by doxycycline in HeLa but not C33A cells. Curiously, when compared to the known Homo sapiens proteome, the representation of tryptophan in the HR-HPV E6 & E6AP degradome is substantially lower, possibly providing a mechanism that underlies the lowered intracellular free tryptophan levels in E6-expressing cells during starvation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

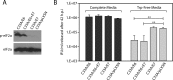
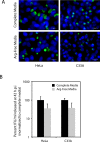
References
-
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282(5389):754–9. Epub 1998/10/23. . - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

