Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana
- PMID: 27658445
- PMCID: PMC5034326
- DOI: 10.1038/srep34027
Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana
Abstract
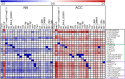
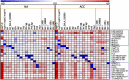
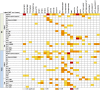
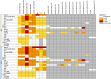
In plants from temperate climates such as Arabidopsis thaliana low, non-freezing temperatures lead to increased freezing tolerance in a process termed cold acclimation. This process is accompanied by massive changes in gene expression and in the content of primary metabolites and lipids. In addition, most flavonols and anthocyanins accumulate upon cold exposure, along with most transcripts encoding transcription factors and enzymes of the flavonoid biosynthetic pathway. However, no evidence for a functional role of flavonoids in plant freezing tolerance has been shown. Here, we present a comprehensive analysis using qRT-PCR for transcript, LC-MS for flavonoid and GC-MS for primary metabolite measurements, and an electrolyte leakage assay to determine freezing tolerance of 20 mutant lines in two Arabidopsis accessions that are affected in different steps of the flavonoid biosynthetic pathway. This analysis provides evidence for a functional role of flavonoids in plant cold acclimation. The accumulation of flavonoids in the activation tagging mutant line pap1-D improved, while reduced flavonoid content in different knock-out mutants impaired leaf freezing tolerance. Analysis of the different knock-out mutants suggests redundancy of flavonoid structures, as the lack of flavonols or anthocyanins could be compensated by other compound classes.
Figures


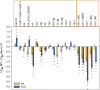
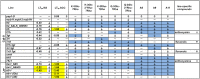
References
-
- Buer C. S., Imin N. & Djordjevic M. A. Flavonoids: new roles for old molecules. J. Integr. Plant Biol. 52, 98–111 (2010). - PubMed
-
- Stobiecki M. A. et al. Profiling of phenolic glycosidic conjugates in leaves of Arabidopsis thaliana using LC/MS. Metabolomics, 197–219 (2006).
-
- Tohge T. et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42, 218–235 (2005). - PubMed
-
- Veit M. & Pauli G. F. Major flavonoids from Arabidopsis thaliana leaves. J. Nat. Prod. 62, 1301–1303 (1999). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

