The Healthy Infant Nasal Transcriptome: A Benchmark Study
- PMID: 27658638
- PMCID: PMC5034274
- DOI: 10.1038/srep33994
The Healthy Infant Nasal Transcriptome: A Benchmark Study
Abstract
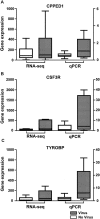
Responses by resident cells are likely to play a key role in determining the severity of respiratory disease. However, sampling of the airways poses a significant challenge, particularly in infants and children. Here, we report a reliable method for obtaining nasal epithelial cell RNA from infants for genome-wide transcriptomic analysis, and describe baseline expression characteristics in an asymptomatic cohort. Nasal epithelial cells were collected by brushing of the inferior turbinates, and gene expression was interrogated by RNA-seq analysis. Reliable recovery of RNA occurred in the absence of adverse events. We observed high expression of epithelial cell markers and similarity to the transcriptome for intrapulmonary airway epithelial cells. We identified genes displaying low and high expression variability, both inherently, and in response to environmental exposures. The greatest gene expression differences in this asymptomatic cohort were associated with the presence of known pathogenic viruses and/or bacteria. Robust bacteria-associated gene expression patterns were significantly associated with the presence of Moraxella. In summary, we have developed a reliable method for interrogating the infant airway transcriptome by sampling the nasal epithelium. Our data demonstrates both the fidelity and feasibility of our methodology, and describes normal gene expression and variation within a healthy infant cohort.
Figures




References
-
- Agusti A., Sobradillo P. & Celli B. Addressing the complexity of chronic obstructive pulmonary disease: from phenotypes and biomarkers to scale-free networks, systems biology, and P4 medicine. American journal of respiratory and critical care medicine 183, 1129–1137 (2011). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

