Overview of the SAMPL5 host-guest challenge: Are we doing better?
- PMID: 27658802
- PMCID: PMC5241188
- DOI: 10.1007/s10822-016-9974-4
Overview of the SAMPL5 host-guest challenge: Are we doing better?
Abstract
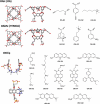
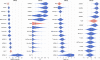
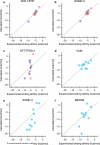
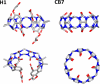
The ability to computationally predict protein-small molecule binding affinities with high accuracy would accelerate drug discovery and reduce its cost by eliminating rounds of trial-and-error synthesis and experimental evaluation of candidate ligands. As academic and industrial groups work toward this capability, there is an ongoing need for datasets that can be used to rigorously test new computational methods. Although protein-ligand data are clearly important for this purpose, their size and complexity make it difficult to obtain well-converged results and to troubleshoot computational methods. Host-guest systems offer a valuable alternative class of test cases, as they exemplify noncovalent molecular recognition but are far smaller and simpler. As a consequence, host-guest systems have been part of the prior two rounds of SAMPL prediction exercises, and they also figure in the present SAMPL5 round. In addition to being blinded, and thus avoiding biases that may arise in retrospective studies, the SAMPL challenges have the merit of focusing multiple researchers on a common set of molecular systems, so that methods may be compared and ideas exchanged. The present paper provides an overview of the host-guest component of SAMPL5, which centers on three different hosts, two octa-acids and a glycoluril-based molecular clip, and two different sets of guest molecules, in aqueous solution. A range of methods were applied, including electronic structure calculations with implicit solvent models; methods that combine empirical force fields with implicit solvent models; and explicit solvent free energy simulations. The most reliable methods tend to fall in the latter class, consistent with results in prior SAMPL rounds, but the level of accuracy is still below that sought for reliable computer-aided drug design. Advances in force field accuracy, modeling of protonation equilibria, electronic structure methods, and solvent models, hold promise for future improvements.
Keywords: Binding affinity; Blind challenge; Computer-aided drug design; Host–guest; Molecular recognition.
Figures


References
-
- Martin E, Ertl P, Hunt P, Duca J, Lewis R. Gazing into the crystal ball; The future of computer-aided drug design. J Comput Aided Mol Des. 2012;26:77–79. doi:10.1007/s10822-011-9487-0. - PubMed
-
- Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–949. doi:10.1038/nrd1549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials

