The genetic and epigenetic landscapes of the epithelium in asthma
- PMID: 27658857
- PMCID: PMC5034566
- DOI: 10.1186/s12931-016-0434-4
The genetic and epigenetic landscapes of the epithelium in asthma
Abstract
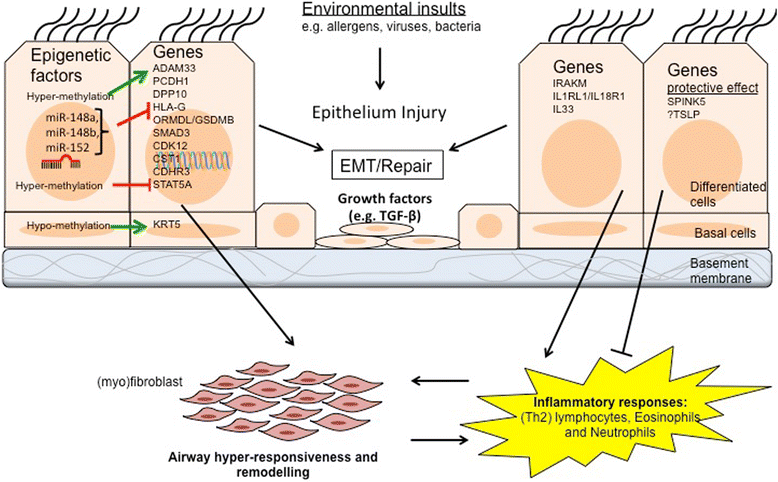
Asthma is a global health problem with increasing prevalence. The airway epithelium is the initial barrier against inhaled noxious agents or aeroallergens. In asthma, the airway epithelium suffers from structural and functional abnormalities and as such, is more susceptible to normally innocuous environmental stimuli. The epithelial structural and functional impairments are now recognised as a significant contributing factor to asthma pathogenesis. Both genetic and environmental risk factors play important roles in the development of asthma with an increasing number of genes associated with asthma susceptibility being expressed in airway epithelium. Epigenetic factors that regulate airway epithelial structure and function are also an attractive area for assessment of susceptibility to asthma. In this review we provide a comprehensive discussion on genetic factors; from using linkage designs and candidate gene association studies to genome-wide association studies and whole genome sequencing, and epigenetic factors; DNA methylation, histone modifications, and non-coding RNAs (especially microRNAs), in airway epithelial cells that are functionally associated with asthma pathogenesis. Our aims were to introduce potential predictors or therapeutic targets for asthma in airway epithelium. Overall, we found very small overlap in asthma susceptibility genes identified with different technologies. Some potential biomarkers are IRAKM, PCDH1, ORMDL3/GSDMB, IL-33, CDHR3 and CST1 in airway epithelial cells. Recent studies on epigenetic regulatory factors have further provided novel insights to the field, particularly their effect on regulation of some of the asthma susceptibility genes (e.g. methylation of ADAM33). Among the epigenetic regulatory mechanisms, microRNA networks have been shown to regulate a major portion of post-transcriptional gene regulation. Particularly, miR-19a may have some therapeutic potential.
Keywords: Asthma; DNA methylation; Epithelial cells; Genes; Histone acetylation; microRNA.
Figures


Similar articles
-
Epigenetic modifying enzyme expression in asthmatic airway epithelial cells and fibroblasts.BMC Pulm Med. 2017 Jan 31;17(1):24. doi: 10.1186/s12890-017-0371-0. BMC Pulm Med. 2017. PMID: 28137284 Free PMC article.
-
Epigenetic Regulation of Airway Epithelium Immune Functions in Asthma.Front Immunol. 2020 Aug 18;11:1747. doi: 10.3389/fimmu.2020.01747. eCollection 2020. Front Immunol. 2020. PMID: 32973742 Free PMC article. Review.
-
Airway epithelial barrier function regulates the pathogenesis of allergic asthma.Clin Exp Allergy. 2014;44(5):620-30. doi: 10.1111/cea.12296. Clin Exp Allergy. 2014. PMID: 24612268 Review.
-
Epithelial cell dysfunction, a major driver of asthma development.Allergy. 2020 Aug;75(8):1902-1917. doi: 10.1111/all.14421. Epub 2020 Jun 16. Allergy. 2020. PMID: 32460363 Free PMC article. Review.
-
Cis- and trans-eQTM analysis reveals novel epigenetic and transcriptomic immune markers of atopic asthma in airway epithelium.J Allergy Clin Immunol. 2023 Oct;152(4):887-898. doi: 10.1016/j.jaci.2023.05.018. Epub 2023 Jun 2. J Allergy Clin Immunol. 2023. PMID: 37271320 Free PMC article.
Cited by
-
Etiology of epithelial barrier dysfunction in patients with type 2 inflammatory diseases.J Allergy Clin Immunol. 2017 Jun;139(6):1752-1761. doi: 10.1016/j.jaci.2017.04.010. J Allergy Clin Immunol. 2017. PMID: 28583447 Free PMC article. Review.
-
The Airway Epithelium-A Central Player in Asthma Pathogenesis.Int J Mol Sci. 2020 Nov 24;21(23):8907. doi: 10.3390/ijms21238907. Int J Mol Sci. 2020. PMID: 33255348 Free PMC article. Review.
-
Sphingosine kinase 1-specific inhibitor PF543 reduces goblet cell metaplasia of bronchial epithelium in an acute asthma model.Am J Physiol Lung Cell Mol Physiol. 2024 Mar 1;326(3):L377-L392. doi: 10.1152/ajplung.00269.2023. Epub 2024 Jan 30. Am J Physiol Lung Cell Mol Physiol. 2024. PMID: 38290992 Free PMC article.
-
CIAPIN1 promotes proliferation and migration of PDGF-BB-activated airway smooth muscle cells via the PI3K/AKT and JAK2/STAT3 signaling pathways.Physiol Rep. 2025 May;13(9):e70360. doi: 10.14814/phy2.70360. Physiol Rep. 2025. PMID: 40338178 Free PMC article.
-
ADAM33's Role in Asthma Pathogenesis: An Overview.Int J Mol Sci. 2024 Feb 15;25(4):2318. doi: 10.3390/ijms25042318. Int J Mol Sci. 2024. PMID: 38396994 Free PMC article. Review.
References
-
- Hackett TL, Singhera GK, Shaheen F, Hayden P, Jackson GR, Hegele RG, Van Eeden S, Bai TR, Dorscheid DR, Knight DA. Intrinsic phenotypic differences of asthmatic epithelium and its inflammatory responses to respiratory syncytial virus and air pollution. Am J Respir Cell Mol Biol. 2011;45(5):1090–1100. doi: 10.1165/rcmb.2011-0031OC. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

