Clinical Genotyping of Non-Small Cell Lung Cancers Using Targeted Next-Generation Sequencing: Utility of Identifying Rare and Co-mutations in Oncogenic Driver Genes
- PMID: 27659017
- PMCID: PMC5031899
- DOI: 10.1016/j.neo.2016.07.010
Clinical Genotyping of Non-Small Cell Lung Cancers Using Targeted Next-Generation Sequencing: Utility of Identifying Rare and Co-mutations in Oncogenic Driver Genes
Abstract
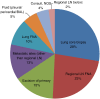
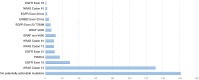
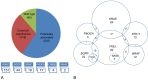
Detection of somatic mutations in non-small cell lung cancers (NSCLCs), especially adenocarcinomas, is important for directing patient care when targeted therapy is available. Here, we present our experience with genotyping NSCLC using the Ion Torrent Personal Genome Machine (PGM) and the AmpliSeq Cancer Hotspot Panel v2. We tested 453 NSCLC samples from 407 individual patients using the 50 gene AmpliSeq Cancer Hotspot Panel v2 from May 2013 to July 2015. Using 10 ng of DNA, up to 11 samples were simultaneously sequenced on the Ion Torrent PGM (316 and 318 chips). We identified variants with the Ion Torrent Variant Caller Plugin, and Golden Helix's SVS software was used for annotation and prediction of the significance of the variants. Three hundred ninety-eight samples were successfully sequenced (12.1% failure rate). In all, 633 variants in 41 genes were detected with a median of 2 (range of 0 to 7) variants per sample. Mutations detected in BRAF, EGFR, ERBB2, KRAS, NRAS, and PIK3CA were considered potentially actionable and were identified in 237 samples, most commonly in KRAS (37.9%), EGFR (11.1%), BRAF (4.8%), and PIK3CA (4.3%). In our patient population, all mutations in EGFR, KRAS, and BRAF were mutually exclusive. The Ion Torrent Ampliseq technology can be utilized on small biopsy and cytology specimens, requires very little input DNA, and can be applied in clinical laboratories for genotyping of NSCLC. This targeted next-generation sequencing approach allows for detection of common and also rare mutations that are clinically actionable in multiple patients simultaneously.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Travis W.D., Brambilla E., Burke A.P., Marx A., Nicholson A.G., editors. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. 4th ed. IAEC; Lyon: 2015. - PubMed
-
- Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013;137:828–860. - PMC - PubMed
-
- National Comprehensive Cancer Network . 2015. NCCN Guidelines Version 4.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

