The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species
- PMID: 27659211
- PMCID: PMC5034548
- DOI: 10.1186/s13059-016-1049-2
The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species
Erratum in
-
Erratum to: The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species.Genome Biol. 2017 Jan 18;18(1):11. doi: 10.1186/s13059-017-1155-9. Genome Biol. 2017. PMID: 28100280 Free PMC article. No abstract available.
Abstract
Background: The Mediterranean fruit fly (medfly), Ceratitis capitata, is a major destructive insect pest due to its broad host range, which includes hundreds of fruits and vegetables. It exhibits a unique ability to invade and adapt to ecological niches throughout tropical and subtropical regions of the world, though medfly infestations have been prevented and controlled by the sterile insect technique (SIT) as part of integrated pest management programs (IPMs). The genetic analysis and manipulation of medfly has been subject to intensive study in an effort to improve SIT efficacy and other aspects of IPM control.
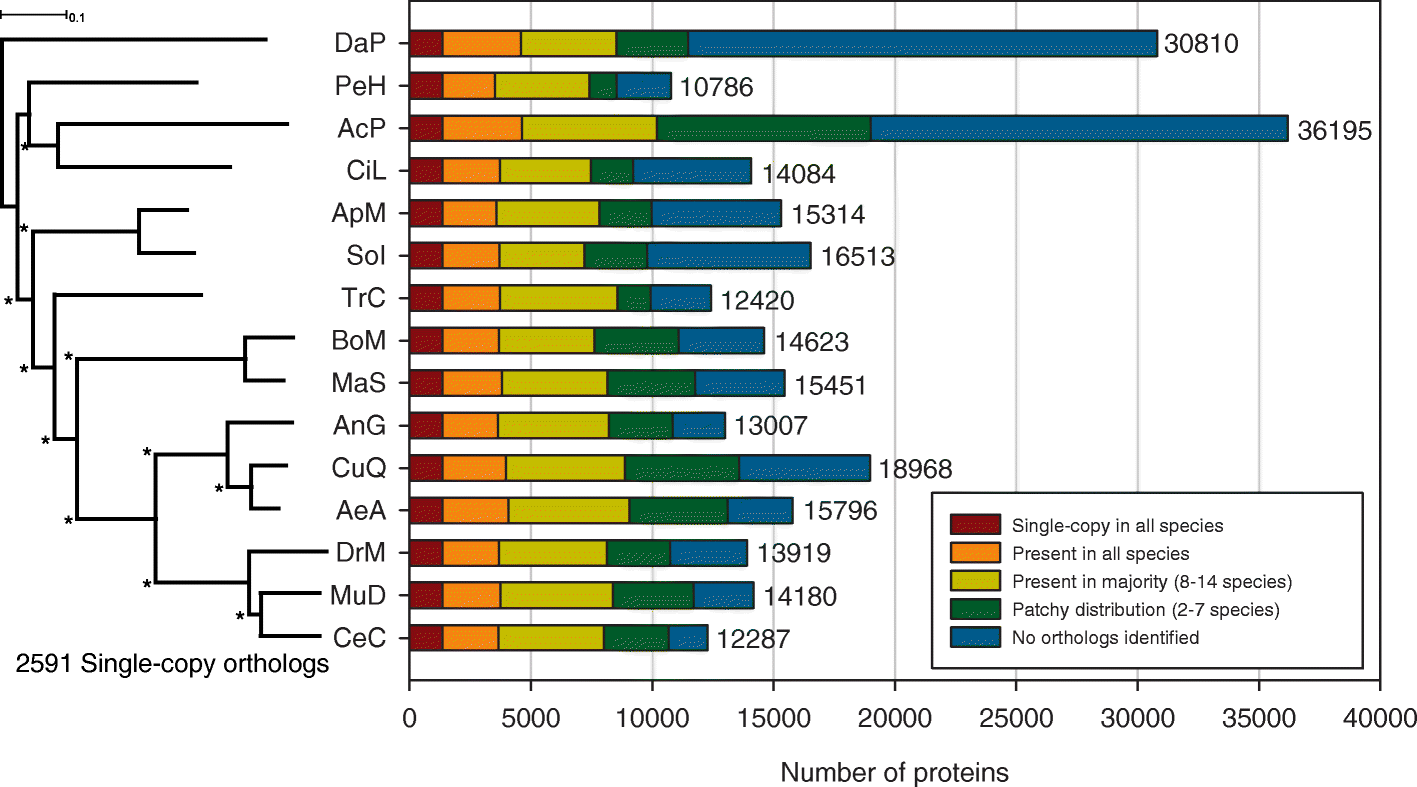
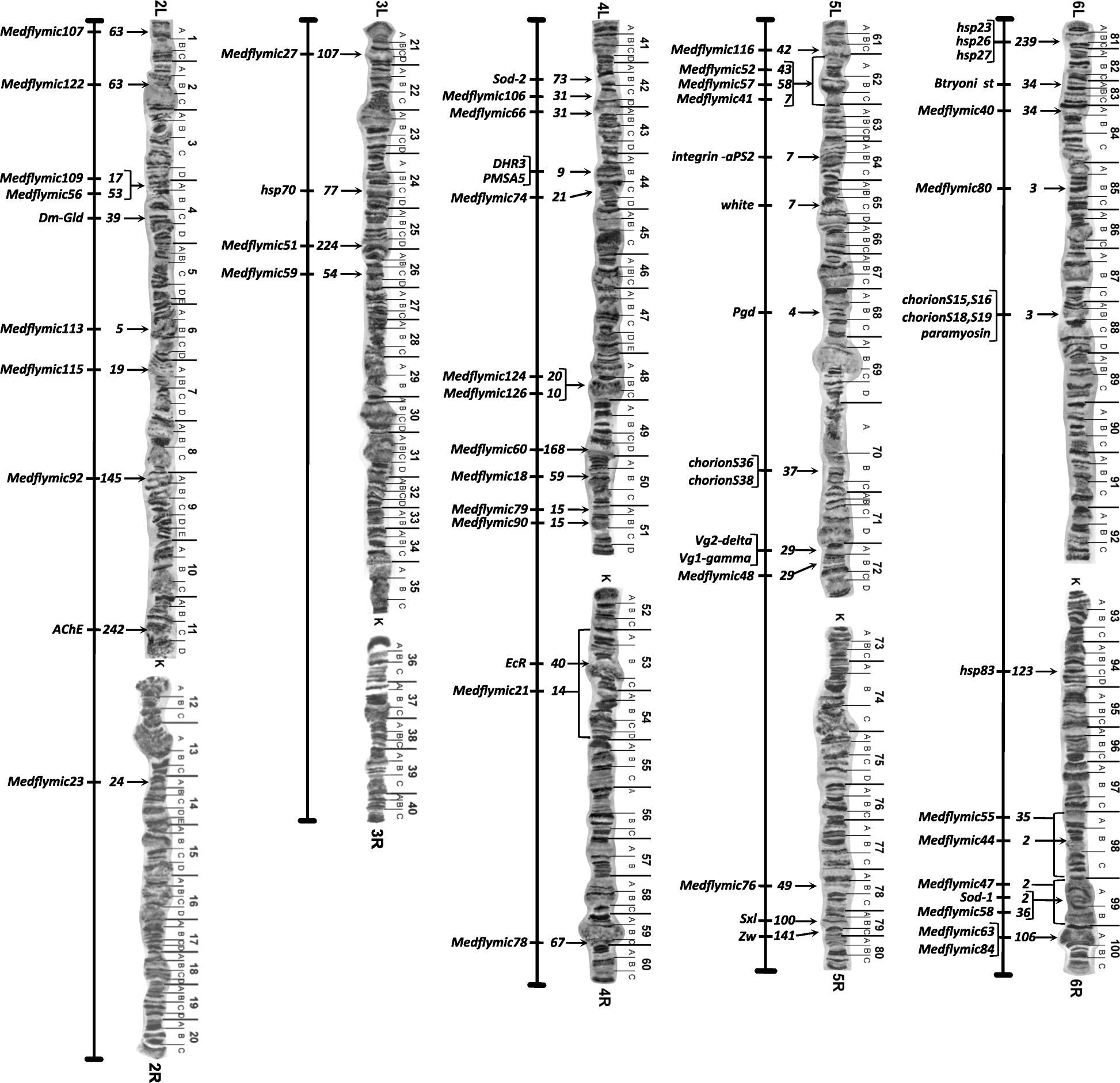
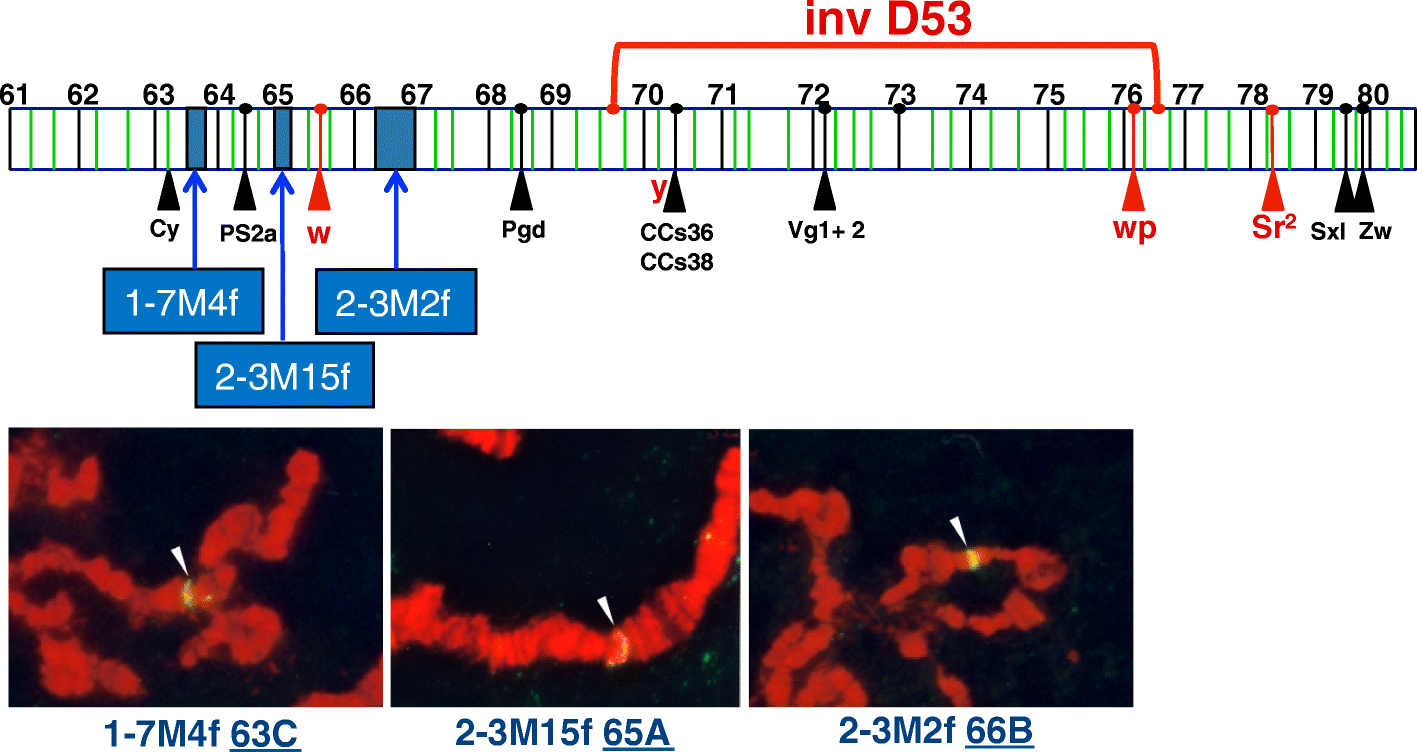
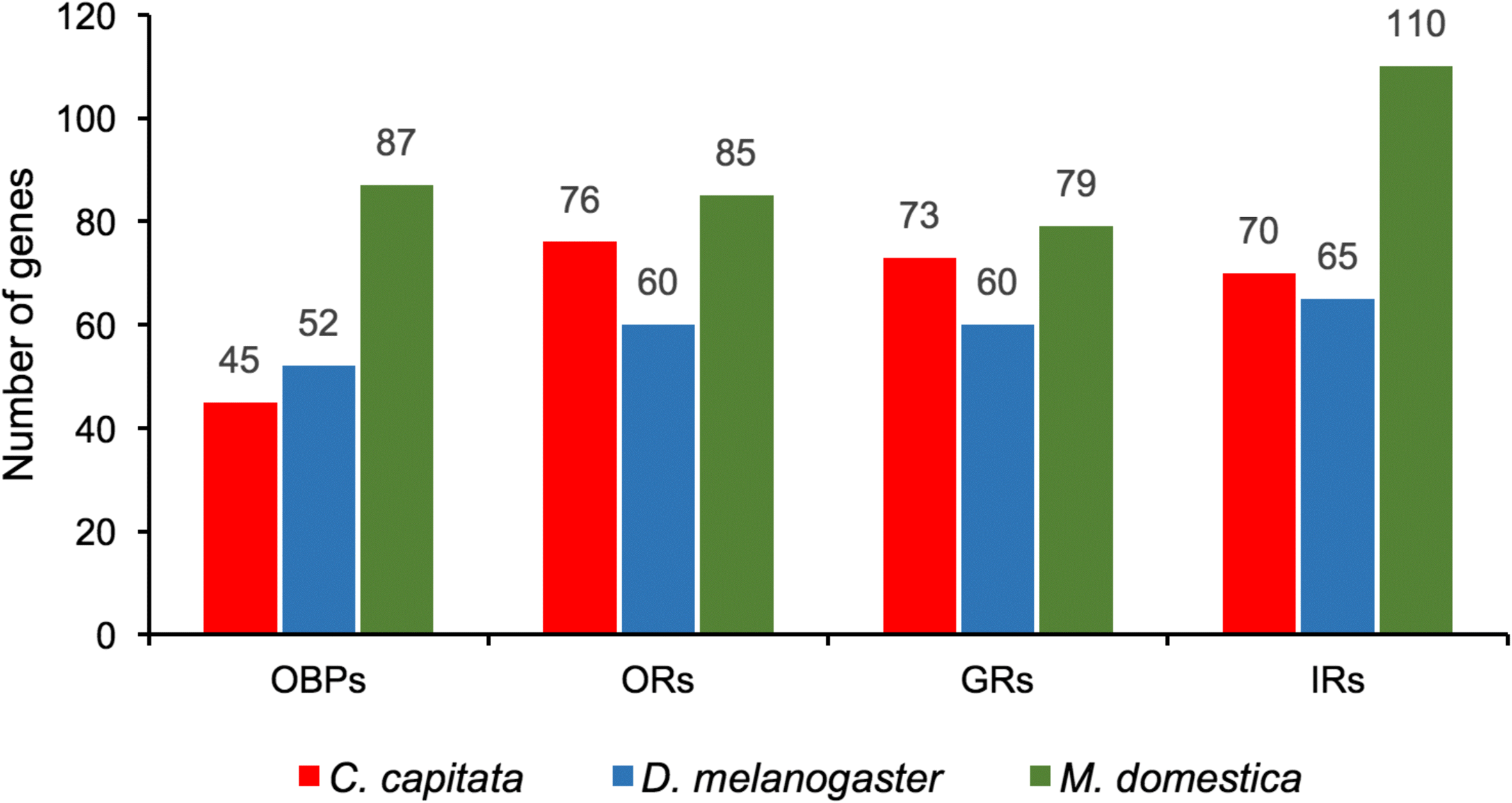
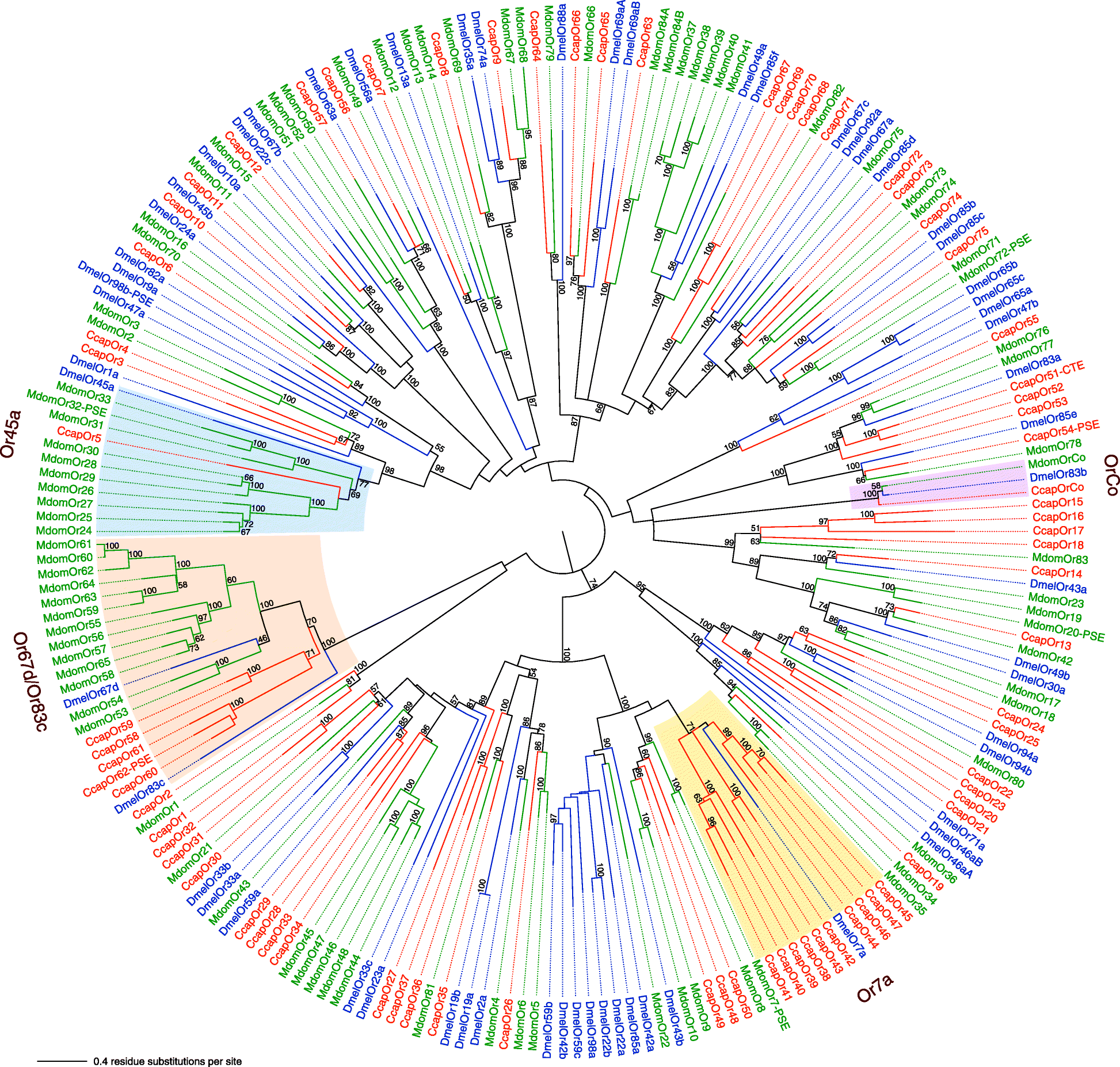
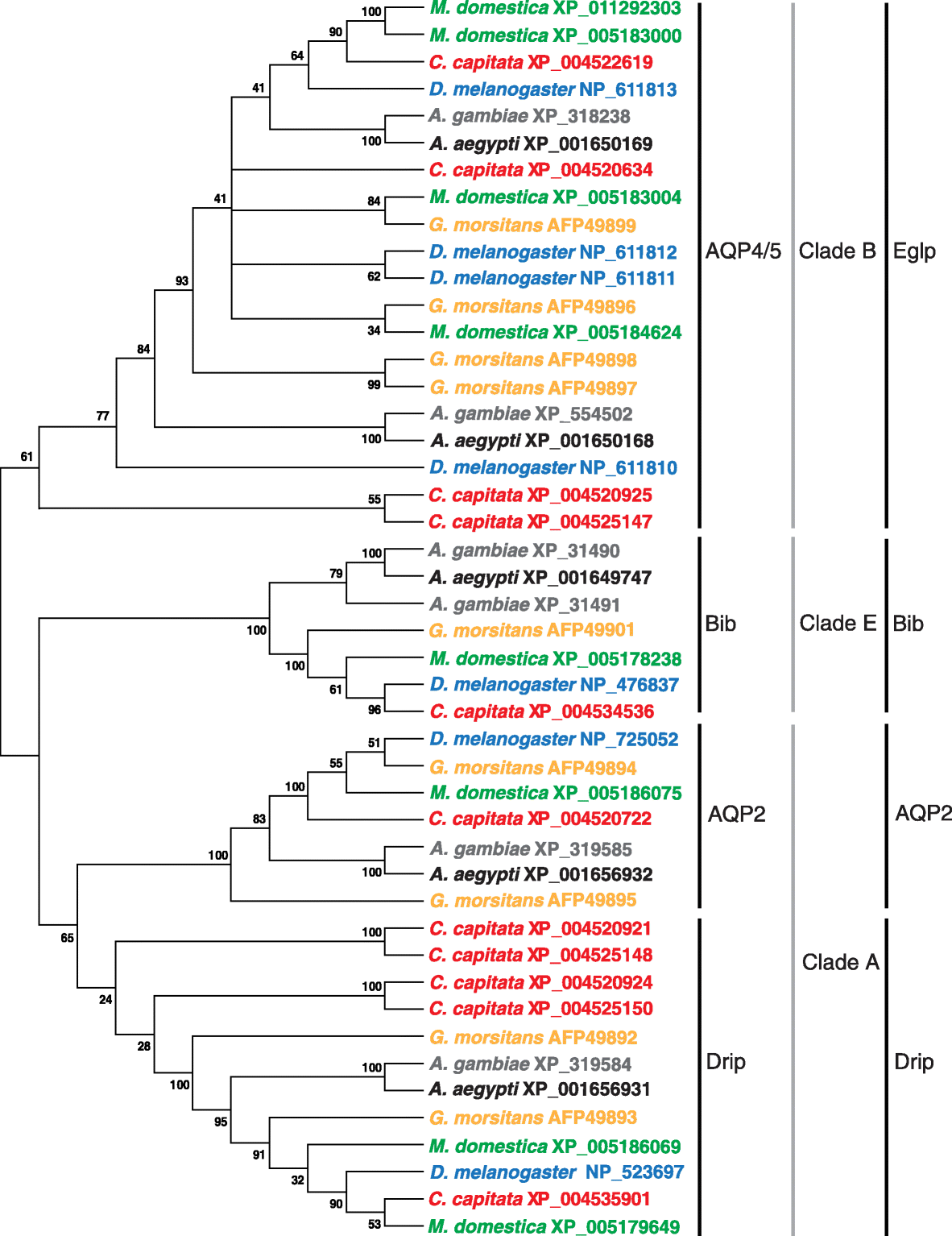
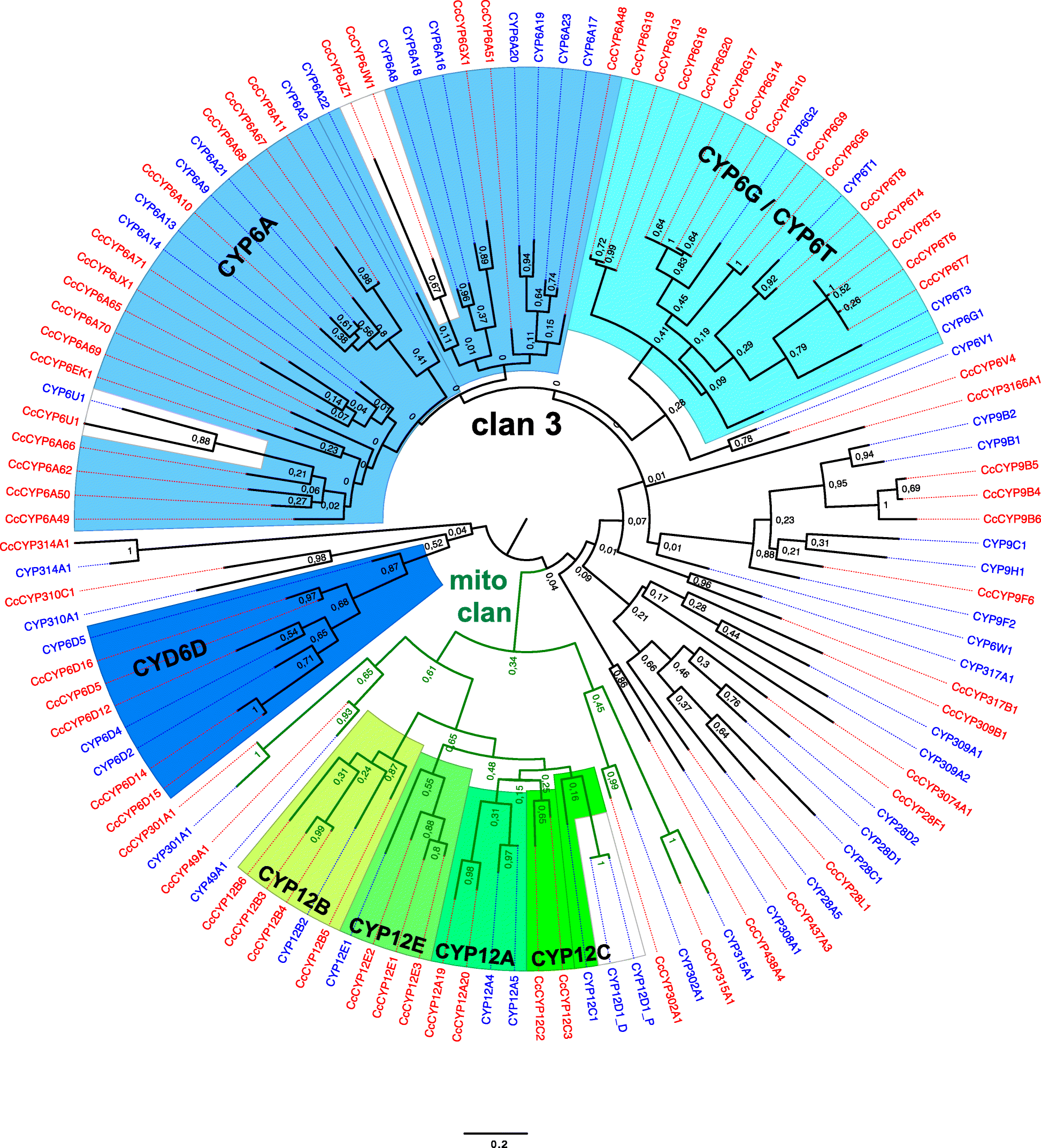
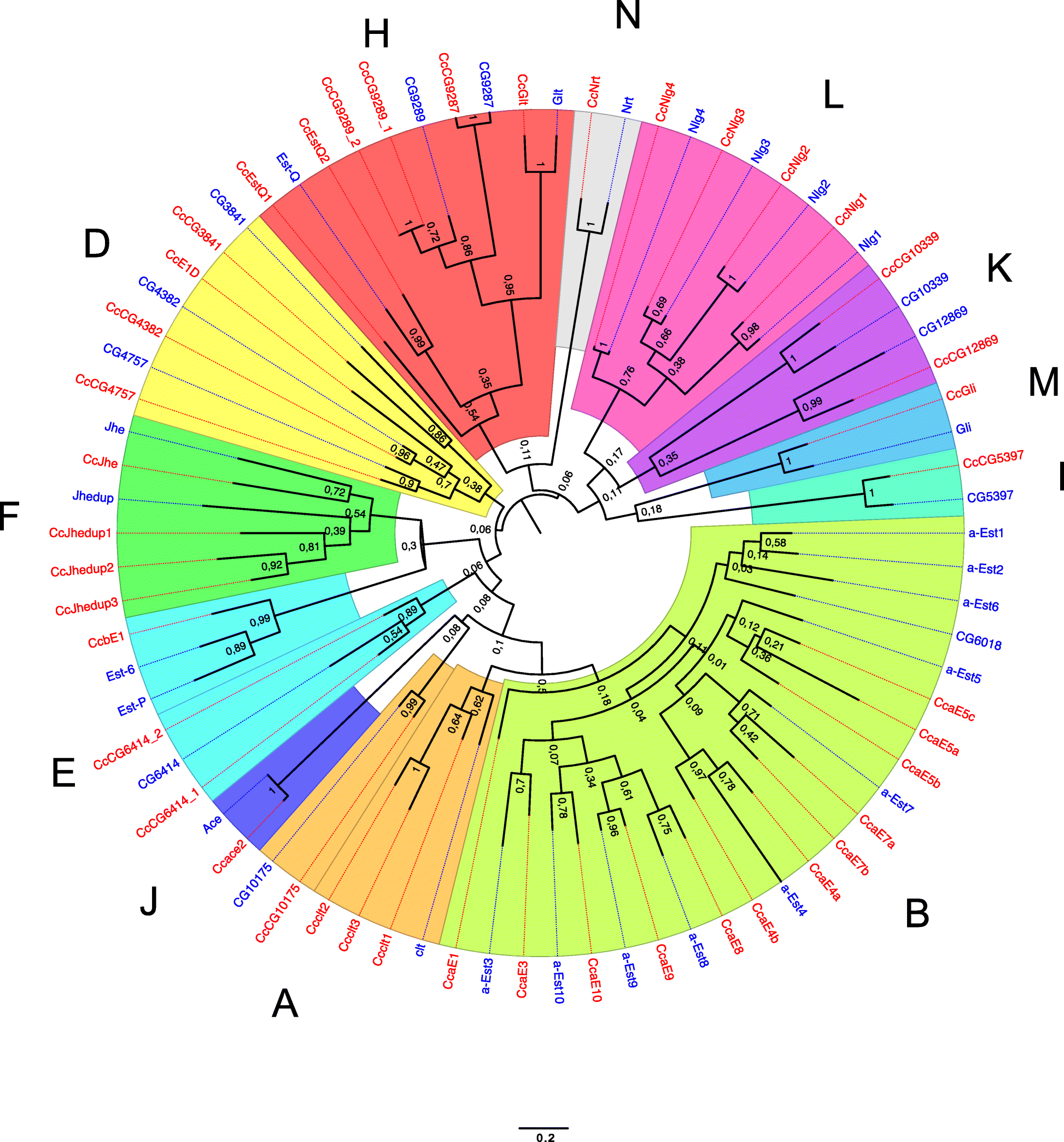
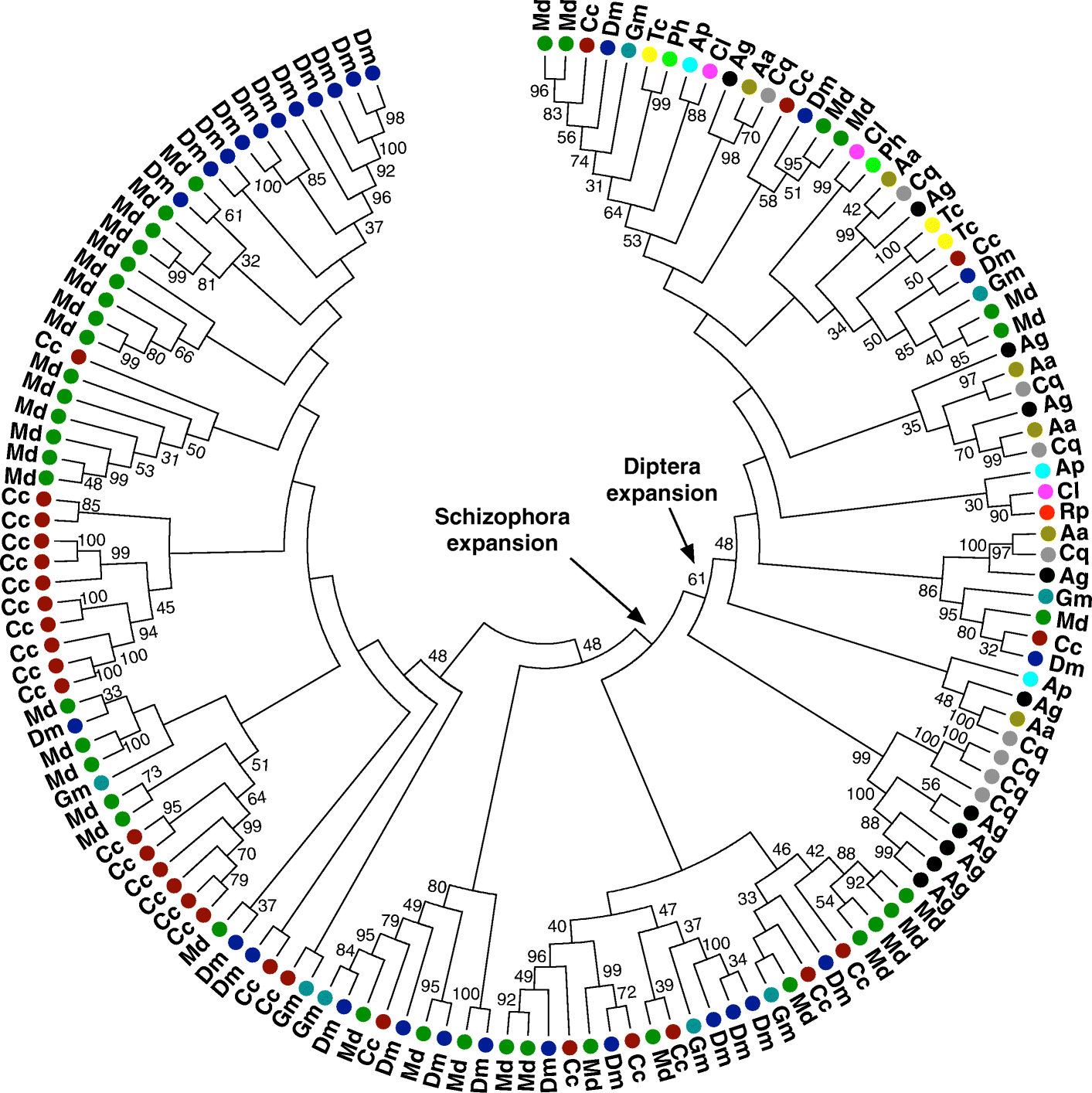
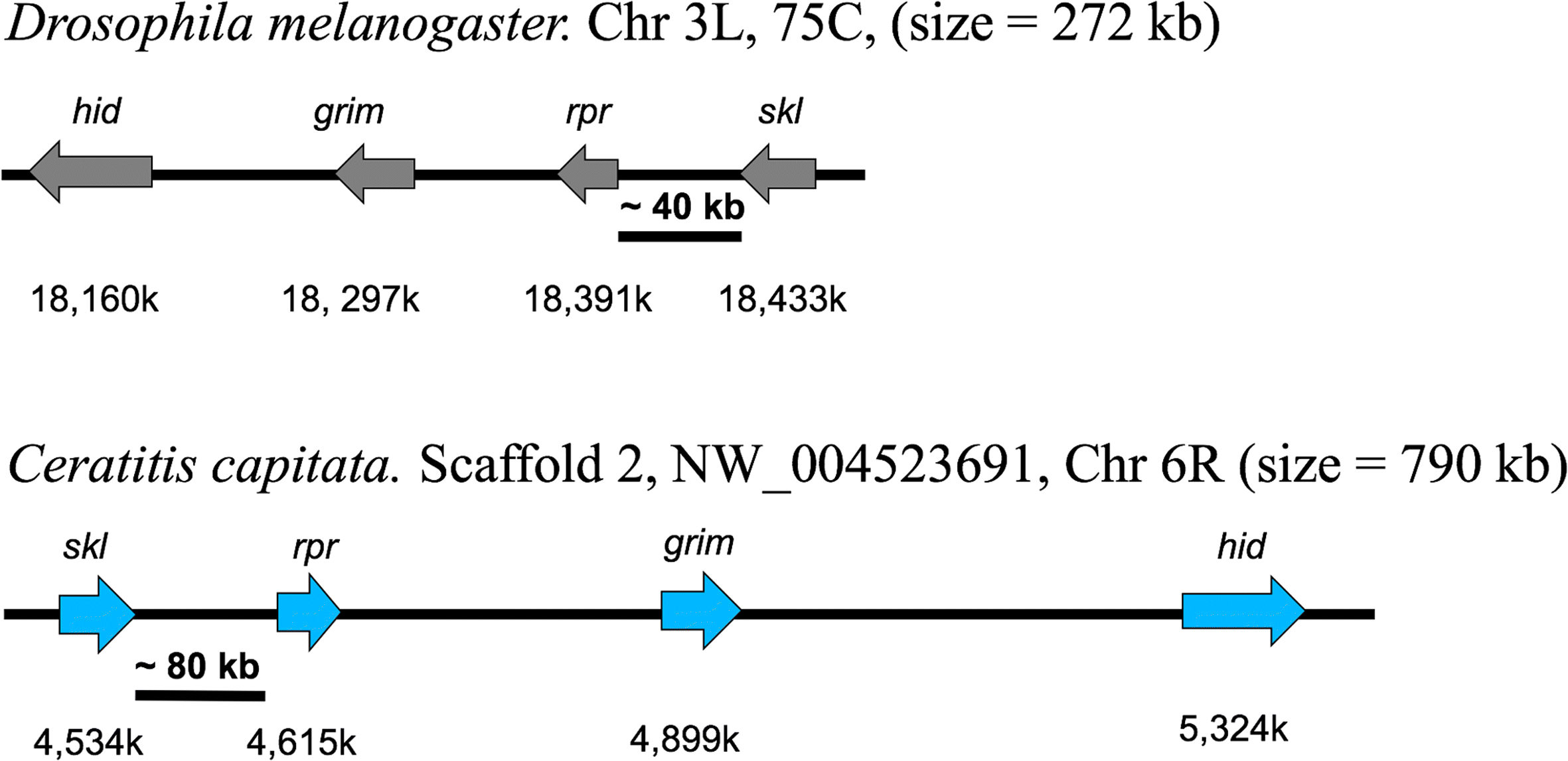
Results: The 479 Mb medfly genome is sequenced from adult flies from lines inbred for 20 generations. A high-quality assembly is achieved having a contig N50 of 45.7 kb and scaffold N50 of 4.06 Mb. In-depth curation of more than 1800 messenger RNAs shows specific gene expansions that can be related to invasiveness and host adaptation, including gene families for chemoreception, toxin and insecticide metabolism, cuticle proteins, opsins, and aquaporins. We identify genes relevant to IPM control, including those required to improve SIT.
Conclusions: The medfly genome sequence provides critical insights into the biology of one of the most serious and widespread agricultural pests. This knowledge should significantly advance the means of controlling the size and invasive potential of medfly populations. Its close relationship to Drosophila, and other insect species important to agriculture and human health, will further comparative functional and structural studies of insect genomes that should broaden our understanding of gene family evolution.
Keywords: Chromosomal synteny; Gene family evolution; Insect adaptation; Insect invasiveness; Insect orthology; Medfly genome; Medfly integrated pest management (IPM); Tephritid genomics.
Figures

References
-
- Liquido NJ, Shinoda LA, Cunningham RT. Host plants of the Mediterranean fruit fly (Diptera, Tephritidae). An annotated world list. Ann Entomol Soc Am. 1991;77:1–57.
-
- Malacrida AR, Guglielmino CR, Gasperi G, Baruffi L, Milani R. Spatial and temporal differentiation in colonizing populations of Ceratitis capitata. Heredity. 1992;69:101–111. doi: 10.1038/hdy.1992.102. - DOI
-
- Enkerlin WR. Impact of fruit fly control programmes using the sterile insect technique. In: Dyck VA, Hendrichs J, Robinson AS, editors. Sterile insect technique - principles and practice in area-wide integrated pest management. Dordrecht: Springer; 2005. pp. 651–676.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

