Activated monocytes resist elimination by retinal pigment epithelium and downregulate their OTX2 expression via TNF-α
- PMID: 27660103
- PMCID: PMC5242302
- DOI: 10.1111/acel.12540
Activated monocytes resist elimination by retinal pigment epithelium and downregulate their OTX2 expression via TNF-α
Abstract
Orthodenticle homeobox 2 (OTX2) controls essential, homeostatic retinal pigment epithelial (RPE) genes in the adult. Using cocultures of human CD14+ blood monocytes (Mos) and primary porcine RPE cells and a fully humanized system using human-induced pluripotent stem cell-derived RPE cells, we show that activated Mos markedly inhibit RPEOTX2 expression and resist elimination in contact with the immunosuppressive RPE. Mechanistically, we demonstrate that TNF-α, secreted from activated Mos, mediates the downregulation of OTX2 and essential RPE genes of the visual cycle among others. Our data show how subretinal, chronic inflammation and in particular TNF-α can affect RPE function, which might contribute to the visual dysfunctions in diseases such as age-related macular degeneration (AMD) where subretinal macrophages are observed. Our findings provide important mechanistic insights into the regulation of OTX2 under inflammatory conditions. Therapeutic restoration of OTX2 expression might help revive RPE and visual function in retinal diseases such as AMD.
Keywords: OTX2; TNF-α; age-related macular degeneration; monocytes; retinal pigment epithelium; visual cycle.
© 2016 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures
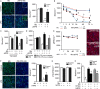
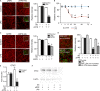
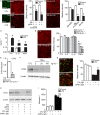

References
-
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brûlet P (1995) Forebrain and midbrain regions are deleted in Otx2‐/‐ mutants due to a defective anterior neuroectoderm specification during gastrulation. Dev. Camb. Engl. 121, 3279–3290. - PubMed
-
- Arnault E, Barrau C, Nanteau C, Gondouin P, Bigot K, Viénot F, Gutman E, Fontaine V, Villette T, Cohen‐Tannoudji D, Sahel J‐A, Picaud S (2013) Phototoxic action spectrum on a retinal pigment epithelium model of age‐related macular degeneration exposed to sunlight normalized conditions. PLoS ONE 8, e71398. - PMC - PubMed
-
- Becroft DM (1981) Prenatal cytomegalovirus infection: epidemiology, pathology and pathogenesis. Perspect. Pediatr. Pathol. 6, 203–241. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

