MicroRNA-552 enhances metastatic capacity of colorectal cancer cells by targeting a disintegrin and metalloprotease 28
- PMID: 27661126
- PMCID: PMC5342546
- DOI: 10.18632/oncotarget.12169
MicroRNA-552 enhances metastatic capacity of colorectal cancer cells by targeting a disintegrin and metalloprotease 28
Abstract
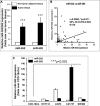
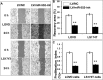
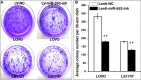
Colorectal cancer (CRC) is one of the most common prevalent cancer types worldwide. MicroRNAs (miRNAs or miRs) have been demonstrated to play crucial roles in the development, metastasis and drug resistance of CRC. In the present study, a strikingly elevated expression of miR-552 was determined in CRC tumor tissues and cells by a miRNA profiling analysis. Importantly, the gene of A Disintegrin And Metalloprotease (ADAM) family member 28 (ADAM28) was identified as a target of miR-552, which was further validated in terms of genetic dual luciferase report assay. Furthermore, an inhibition of miR-552 in LOVE and LS174T CRC cells by transducing miR-552 inhibitor (antagomiR-552) with a lentiviral vector exhibited an ability to reduce cell proliferation, migration and clonogenicity. Moreover, both LOVO and LS174T cells stably expressing miR-552 inhibitor displayed a decreased ability to develop tumors in a murine xenograft model in vivo. In contrast, a knockdown of ADAM28 by short hairpin RNA could reverse the antagomiR-552-induced inhibition of metastatic features of CRC cells in vitro. These results suggested that miR-552 is an oncomir able to promote CRC metastasis in part through a mechanism of targeting ADAM28, which may be a novel target for CRC treatment and warrants for further investigation.
Keywords: ADAM28; antagomir; colorectal cancer; miR-552; microRNAs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. - PubMed
-
- Cojoc M, Mabert K, Muders MH, Dubrovska A. A role for cancer stem cells in therapy resistance: cellular and molecular mechanisms. Seminars in cancer biology. 2015;31:16–27. - PubMed
-
- Kartal-Yandim M, Adan-Gokbulut A, Baran Y. Molecular mechanisms of drug resistance and its reversal in cancer. Critical reviews in biotechnology. 2015:1–11. - PubMed
-
- Lippert TH, Ruoff HJ, Volm M. Intrinsic and acquired drug resistance in malignant tumors. The main reason for therapeutic failure. Arzneimittel-Forschung. 2008;58:261–264. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

