Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation
- PMID: 27661255
- PMCID: PMC5094855
- DOI: 10.7554/eLife.19760
Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation
Abstract
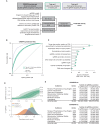
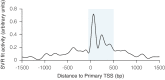
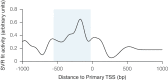
We recently found that nucleosomes directly block access of CRISPR/Cas9 to DNA (Horlbeck et al., 2016). Here, we build on this observation with a comprehensive algorithm that incorporates chromatin, position, and sequence features to accurately predict highly effective single guide RNAs (sgRNAs) for targeting nuclease-dead Cas9-mediated transcriptional repression (CRISPRi) and activation (CRISPRa). We use this algorithm to design next-generation genome-scale CRISPRi and CRISPRa libraries targeting human and mouse genomes. A CRISPRi screen for essential genes in K562 cells demonstrates that the large majority of sgRNAs are highly active. We also find CRISPRi does not exhibit any detectable non-specific toxicity recently observed with CRISPR nuclease approaches. Precision-recall analysis shows that we detect over 90% of essential genes with minimal false positives using a compact 5 sgRNA/gene library. Our results establish CRISPRi and CRISPRa as premier tools for loss- or gain-of-function studies and provide a general strategy for identifying Cas9 target sites.
Keywords: CRISPR; chromosomes; computational biology; genes; genetic screening; human; nucleosomes; systems biology.
Conflict of interest statement
MAH: patent application related to CRISPRi and CRISPRa screening (PCT/US15/40449). JSW is a founder of, and MAH and LAG are consultants for, KSQ Therapeutics, a CRISPR functional genomics company. LAG: filed a patent application related to CRISPRi and CRISPRa screening (PCT/US15/40449). JSW is a founder of, and MAH and LAG are consultants for, KSQ Therapeutics, a CRISPR functional genomics company. MK: patent application related to CRISPRi and CRISPRa screening (PCT/US15/40449). JSW: filed a patent application related to CRISPRi and CRISPRa screening (PCT/US15/40449). JSW is a founder of, and MAH and LAG are consultants for, KSQ Therapeutics, a CRISPR functional genomics company. The other authors declare that no competing interests exist.
Figures






Comment in
-
Making better CRISPR libraries.Elife. 2016 Nov 3;5:e21863. doi: 10.7554/eLife.21863. Elife. 2016. PMID: 27809196 Free PMC article.
References
-
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, Kost-Alimova M, Gill S, Xu H, Ali LD, Jiang G, Pantel S, Lee Y, Goodale A, Cherniack AD, Oh C, Kryukov G, Cowley GS, Garraway LA, Stegmaier K, Roberts CW, Golub TR, Meyerson M, Root DE, Tsherniak A, Hahn WC. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discovery. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. - DOI - PMC - PubMed
-
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, Hyman AA, Leproust EM, McManus MT, Weissman JS. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. - DOI - PMC - PubMed
-
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM, Iyer PR, Lin E, Guzman S. Highly efficient Cas9-mediated transcriptional programming. Nature Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

