Escape to Ferality: The Endoferal Origin of Weedy Rice from Crop Rice through De-Domestication
- PMID: 27661982
- PMCID: PMC5035073
- DOI: 10.1371/journal.pone.0162676
Escape to Ferality: The Endoferal Origin of Weedy Rice from Crop Rice through De-Domestication
Abstract
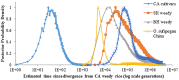
Domestication is the hallmark of evolution and civilization and harnesses biodiversity through selection for specific traits. In regions where domesticated lines are grown near wild relatives, congeneric sources of aggressive weedy genotypes cause major economic losses. Thus, the origins of weedy genotypes where no congeneric species occur raise questions regarding management effectiveness and evolutionary mechanisms responsible for weedy population success. Since eradication in the 1970s, California growers avoided weedy rice through continuous flood culture and zero-tolerance guidelines, preventing the import, presence, and movement of weedy seeds. In 2003, after decades of no reported presence in California, a weedy rice population was confirmed in dry-seeded fields. Our objectives were to identify the origins and establishment of this population and pinpoint possible phenotypes involved. We show that California weedy rice is derived from a different genetic source among a broad range of AA genome Oryzas and is most recently diverged from O. sativa temperate japonica cultivated in California. In contrast, other weedy rice ecotypes in North America (Southern US) originate from weedy genotypes from China near wild Oryza, and are derived through existing crop-wild relative crosses. Analyses of morphological data show that California weedy rice subgroups have phenotypes like medium-grain or gourmet cultivars, but have colored pericarp, seed shattering, and awns like wild relatives, suggesting that reversion to non-domestic or wild-like traits can occur following domestication, despite apparent fixation of domestication alleles. Additionally, these results indicate that preventive methods focused on incoming weed sources through contamination may miss burgeoning weedy genotypes that rapidly adapt, establish, and proliferate. Investigating the common and unique evolutionary mechanisms underlying global weed origins and subsequent interactions with crop relatives sheds light on how weeds evolve and addresses broader questions regarding the stability of selection during domestication and crop improvement.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
African Cultivated, Wild and Weedy Rice (Oryza spp.): Anticipating Further Genomic Studies.Biology (Basel). 2024 Sep 5;13(9):697. doi: 10.3390/biology13090697. Biology (Basel). 2024. PMID: 39336124 Free PMC article. Review.
-
Contrasting patterns of variation in weedy traits and unique crop features in divergent populations of US weedy rice (Oryza sativa sp.) in Arkansas and California.Pest Manag Sci. 2018 Jun;74(6):1404-1415. doi: 10.1002/ps.4820. Epub 2018 Mar 5. Pest Manag Sci. 2018. PMID: 29205860
-
Malaysian weedy rice shows its true stripes: wild Oryza and elite rice cultivars shape agricultural weed evolution in Southeast Asia.Mol Ecol. 2014 Oct;23(20):5003-17. doi: 10.1111/mec.12922. Epub 2014 Oct 13. Mol Ecol. 2014. PMID: 25231087
-
The Role of Standing Variation in the Evolution of Weedines Traits in South Asian Weedy Rice (Oryza spp.).G3 (Bethesda). 2018 Nov 6;8(11):3679-3690. doi: 10.1534/g3.118.200605. G3 (Bethesda). 2018. PMID: 30275171 Free PMC article.
-
Genetic Diversity of Weedy Rice and Its Potential Application as a Novel Source of Disease Resistance.Plants (Basel). 2023 Aug 2;12(15):2850. doi: 10.3390/plants12152850. Plants (Basel). 2023. PMID: 37571004 Free PMC article. Review.
Cited by
-
Engineering Herbicide-Tolerance Rice Expressing an Acetohydroxyacid Synthase with a Single Amino Acid Deletion.Int J Mol Sci. 2020 Feb 13;21(4):1265. doi: 10.3390/ijms21041265. Int J Mol Sci. 2020. PMID: 32070060 Free PMC article.
-
Comparative Mapping of Seed Dormancy Loci Between Tropical and Temperate Ecotypes of Weedy Rice (Oryza sativa L.).G3 (Bethesda). 2017 Aug 7;7(8):2605-2614. doi: 10.1534/g3.117.040451. G3 (Bethesda). 2017. PMID: 28592557 Free PMC article.
-
African Cultivated, Wild and Weedy Rice (Oryza spp.): Anticipating Further Genomic Studies.Biology (Basel). 2024 Sep 5;13(9):697. doi: 10.3390/biology13090697. Biology (Basel). 2024. PMID: 39336124 Free PMC article. Review.
-
Becoming weeds.Nat Genet. 2017 Apr 26;49(5):654-655. doi: 10.1038/ng.3851. Nat Genet. 2017. PMID: 28442795
-
Call of the wild rice: Oryza rufipogon shapes weedy rice evolution in Southeast Asia.Evol Appl. 2018 Jan 11;12(1):93-104. doi: 10.1111/eva.12581. eCollection 2019 Jan. Evol Appl. 2018. PMID: 30622638 Free PMC article.
References
-
- Ellstrand NC, Prentice HC, Hancock JF. Gene flow and introgression from domesticated plants into their wild relatives. Annual Review of Ecology and Systematics. 1999;30: 539–563.
-
- Dobney K, Larson G. Genetics and animal domestication: new windows on an elusive process. J Zool. 2006;269: 261–271.
-
- Lawton-Rauh A, Robichaux RH, Purugganan MD. Diversity and divergence patterns in regulatory genes suggest differential gene flow in recently-derived species of the Hawaiian silversword alliance adaptive radiation (Heliantheae, Asteraceae). Mol Ecol. 2007;16: 3995–4013. - PubMed

