Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing
- PMID: 27662090
- PMCID: PMC5039111
- DOI: 10.1016/j.cell.2016.09.006
Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing
Abstract
Gene silencing is instrumental to interrogate gene function and holds promise for therapeutic applications. Here, we repurpose the endogenous retroviruses' silencing machinery of embryonic stem cells to stably silence three highly expressed genes in somatic cells by epigenetics. This was achieved by transiently expressing combinations of engineered transcriptional repressors that bind to and synergize at the target locus to instruct repressive histone marks and de novo DNA methylation, thus ensuring long-term memory of the repressive epigenetic state. Silencing was highly specific, as shown by genome-wide analyses, sharply confined to the targeted locus without spreading to nearby genes, resistant to activation induced by cytokine stimulation, and relieved only by targeted DNA demethylation. We demonstrate the portability of this technology by multiplex gene silencing, adopting different DNA binding platforms and interrogating thousands of genomic loci in different cell types, including primary T lymphocytes. Targeted epigenome editing might have broad application in research and medicine.
Keywords: B2M-null cells; CRISPR/Cas9; DNA methylation; DNMT3L; KRAB-ZFP/KAP1; TALE; TET1; epigenetic editing; gene therapy; permanent gene silencing.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

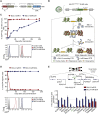

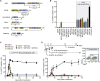




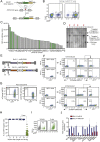
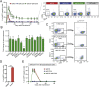
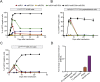

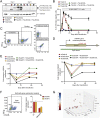
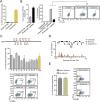

References
-
- Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

