A Broad Set of Chromatin Factors Influences Splicing
- PMID: 27662573
- PMCID: PMC5035054
- DOI: 10.1371/journal.pgen.1006318
A Broad Set of Chromatin Factors Influences Splicing
Abstract
Several studies propose an influence of chromatin on pre-mRNA splicing, but it is still unclear how widespread and how direct this phenomenon is. We find here that when assembled in vivo, the U2 snRNP co-purifies with a subset of chromatin-proteins, including histones and remodeling complexes like SWI/SNF. Yet, an unbiased RNAi screen revealed that the outcome of splicing is influenced by a much larger variety of chromatin factors not all associating with the spliceosome. The availability of this broad range of chromatin factors impacting splicing further unveiled their very context specific effect, resulting in either inclusion or skipping, depending on the exon under scrutiny. Finally, a direct assessment of the impact of chromatin on splicing using an in vitro co-transcriptional splicing assay with pre-mRNAs transcribed from a nucleosomal template, demonstrated that chromatin impacts nascent pre-mRNP in their competence for splicing. Altogether, our data show that numerous chromatin factors associated or not with the spliceosome can affect the outcome of splicing, possibly as a function of the local chromatin environment that by default interferes with the efficiency of splicing.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
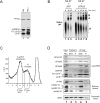


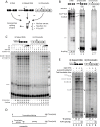
References
-
- Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. Reik W, editor. PLoS Genet. Public Library of Science; 2012;8: e1002717 10.1371/journal.pgen.1002717 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

