Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers
- PMID: 27663357
- PMCID: PMC5034533
- DOI: 10.1186/s12885-016-2785-0
Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers
Abstract
Background: Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous group of tumours with a typical 5 year survival rate of <40 %. DNA methylation in tumour-suppressor genes often occurs at an early stage of tumorigenesis, hence DNA methylation can be used as an early tumour biomarker. Saliva is an ideal diagnostic medium to detect early HNSCC tumour activities due to its proximity to tumour site, non-invasiveness and ease of sampling. We test the hypothesis that the surveillance of DNA methylation in five tumour-suppressor genes (RASSF1α, p16 INK4a , TIMP3, PCQAP/MED15) will allow us to diagnose HNSCC patients from a normal healthy control group as well as to discriminate between Human Papillomavirus (HPV)-positive and HPV-negative patients.
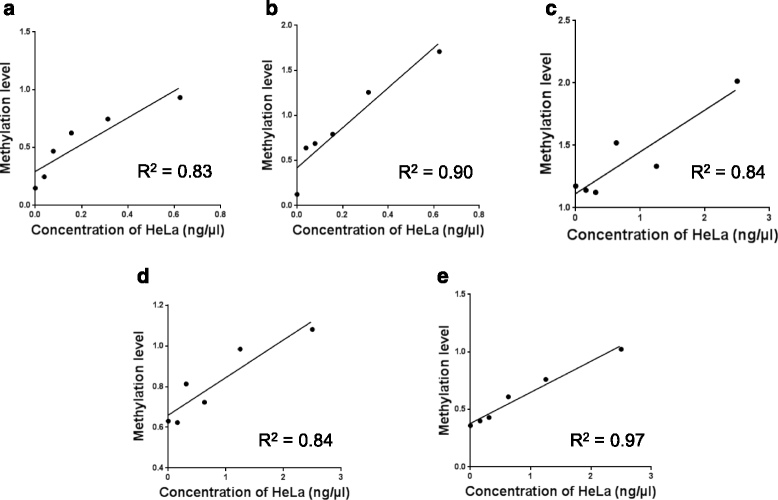
Methods: Methylation-specific PCR (MSP) was used to determine the methylation levels of RASSF1α, p16 INK4a , TIMP3 and PCQAP/MED15 in DNA isolated from saliva. Statistical analysis was carried out using non-parametric Mann-Whitney's U-test for individually methylated genes. A logistic regression analysis was carried out to determine the assay sensitivity when combing the five genes. Further, a five-fold cross-validation with a bootstrap procedure was carried out to determine how well the panel will perform in a real clinical scenario.
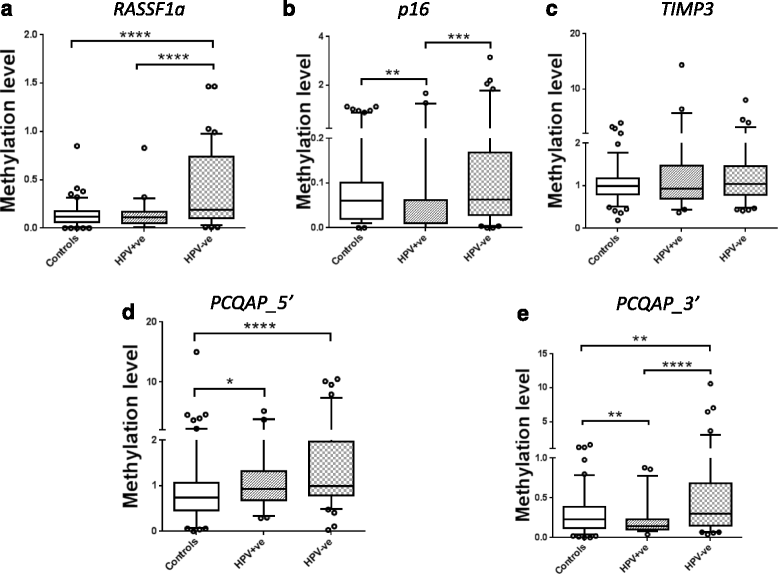
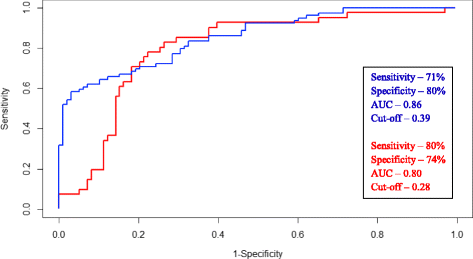
Results: Salivary DNA methylation levels were not affected by age. Salivary DNA methylation levels for RASSF1α, p16 INK4a , TIMP3 and PCQAP/MED15 were higher in HPV-negative HNSCC patients (n = 88) compared with a normal healthy control group (n = 122) (sensitivity of 71 % and specificity of 80 %). Conversely, DNA methylation levels for these genes were lower in HPV-positive HNSCC patients (n = 45) compared with a normal healthy control group (sensitivity of 80 % and specificity of 74 %), consistent with the proposed aetiology of HPV-positive HNSCCs.
Conclusions: Salivary DNA tumour-suppressor methylation gene panel has the potential to detect early-stage tumours in HPV-negative HNSCC patients. HPV infection was found to deregulate the methylation levels in HPV-positive HNSCC patients. Large-scale double-blinded clinical trials are crucial before this panel can potentially be integrated into a clinical setting.
Keywords: Cross-fold validation and early detection; DNA methylation; Epigenetics biomarkers; Head and neck cancers; Human Papillomavirus; Saliva; Tumour-suppressor genes.
Figures
Similar articles
-
Promoter Hypermethylation of Tumor-Suppressor Genes p16INK4a,RASSF1A,TIMP3, and PCQAP/MED15 in Salivary DNA as a Quadruple Biomarker Panel for Early Detection of Oral and Oropharyngeal Cancers.Biomolecules. 2019 Apr 12;9(4):148. doi: 10.3390/biom9040148. Biomolecules. 2019. PMID: 31013839 Free PMC article.
-
DNA Methylation at the Novel CpG Sites in the Promoter of MED15/PCQAP Gene as a Biomarker for Head and Neck Cancers.Biomark Insights. 2014 Jul 3;9:53-60. doi: 10.4137/BMI.S16199. eCollection 2014. Biomark Insights. 2014. PMID: 25057238 Free PMC article.
-
Salivary miRNA panel to detect HPV-positive and HPV-negative head and neck cancer patients.Oncotarget. 2017 Oct 10;8(59):99990-100001. doi: 10.18632/oncotarget.21725. eCollection 2017 Nov 21. Oncotarget. 2017. PMID: 29245955 Free PMC article.
-
MicroRNAs as new biomarkers for human papilloma virus related head and neck cancers.Cancer Biomark. 2015;15(3):213-8. doi: 10.3233/CBM-150464. Cancer Biomark. 2015. PMID: 25769448 Review.
-
DNA Methylation and HPV-Associated Head and Neck Cancer.Microorganisms. 2021 Apr 10;9(4):801. doi: 10.3390/microorganisms9040801. Microorganisms. 2021. PMID: 33920277 Free PMC article. Review.
Cited by
-
Methylation silencing of TGF-β receptor type II is involved in malignant transformation of esophageal squamous cell carcinoma.Clin Epigenetics. 2020 Feb 11;12(1):25. doi: 10.1186/s13148-020-0819-6. Clin Epigenetics. 2020. PMID: 32046777 Free PMC article.
-
Analysis of salivary detection of P16INK4A and RASSF1A promoter gene methylation and its association with oral squamous cell carcinoma in a Colombian population.J Clin Exp Dent. 2020 May 1;12(5):e452-e460. doi: 10.4317/jced.56647. eCollection 2020 May. J Clin Exp Dent. 2020. PMID: 32509227 Free PMC article.
-
Research progress and applications of epigenetic biomarkers in cancer.Front Pharmacol. 2024 Apr 12;15:1308309. doi: 10.3389/fphar.2024.1308309. eCollection 2024. Front Pharmacol. 2024. PMID: 38681199 Free PMC article. Review.
-
Head and neck cancer patient-derived tumouroid cultures: opportunities and challenges.Br J Cancer. 2023 May;128(10):1807-1818. doi: 10.1038/s41416-023-02167-4. Epub 2023 Feb 10. Br J Cancer. 2023. PMID: 36765173 Free PMC article. Review.
-
Cancer Epigenetic Biomarkers in Liquid Biopsy for High Incidence Malignancies.Cancers (Basel). 2021 Jun 16;13(12):3016. doi: 10.3390/cancers13123016. Cancers (Basel). 2021. PMID: 34208598 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

