Prognostic Utility of a New mRNA Expression Signature of Gleason Score
- PMID: 27663590
- PMCID: PMC5215643
- DOI: 10.1158/1078-0432.CCR-16-1245
Prognostic Utility of a New mRNA Expression Signature of Gleason Score
Abstract
Purpose: Gleason score strongly predicts prostate cancer mortality; however, scoring varies among pathologists, and many men are diagnosed with intermediate-risk Gleason score 7. We previously developed a 157-gene signature for Gleason score using a limited gene panel. Using a new whole-transcriptome expression dataset, we verified the previous signature's performance and developed a new Gleason signature to improve lethal outcome prediction among men with Gleason score 7.
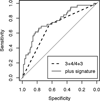
Experimental design: We generated mRNA expression data from prostate tumor tissue from men in the Physicians' Health Study and Health Professionals Follow-Up Study (N = 404) using the Affymetrix Human Gene 1.0 ST microarray. The Prediction Analysis for Microarrays method was used to develop a signature to distinguish high (≥8) versus low (≤6) Gleason score. We evaluated the signature's ability to improve prediction of lethality among men with Gleason score 7, adjusting for 3 + 4/4 + 3 status, by quantifying the area under the receiver operating characteristic (ROC) curve (AUC).
Results: We identified a 30-gene signature that best distinguished Gleason score ≤6 from ≥8. The AUC to predict lethal disease among Gleason score 7 men was 0.76 [95% confidence interval (CI), 0.67-0.84] compared with 0.68 (95% CI, 0.59-0.76) using 3 + 4/4 + 3 status alone (P = 0.0001). This signature was a nonsignificant (P = 0.09) improvement over our previous signature (AUC = 0.72).
Conclusions: Our new 30-gene signature improved prediction of lethality among men with Gleason score 7. This signature can potentially become a useful prognostic tool for physicians to improve treatment decision making. Clin Cancer Res; 23(1); 81-87. ©2016 AACRSee related commentary by Yin et al., p. 6.
©2016 American Association for Cancer Research.
Conflict of interest statement
We have no conflicts of interest to disclose.
Figures


References
MeSH terms
Grants and funding
- P50 CA090381/CA/NCI NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- R01 HL034595/HL/NHLBI NIH HHS/United States
- R01 CA131945/CA/NCI NIH HHS/United States
- R01 CA133891/CA/NCI NIH HHS/United States
- R01 HL026490/HL/NHLBI NIH HHS/United States
- R01 CA097193/CA/NCI NIH HHS/United States
- T32 CA009001/CA/NCI NIH HHS/United States
- UM1 CA167552/CA/NCI NIH HHS/United States
- R01 CA040360/CA/NCI NIH HHS/United States
- R01 CA136578/CA/NCI NIH HHS/United States
- R01 CA034944/CA/NCI NIH HHS/United States
- R01 CA141298/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

