Evolution of Cilia
- PMID: 27663773
- PMCID: PMC5204320
- DOI: 10.1101/cshperspect.a028290
Evolution of Cilia
Abstract
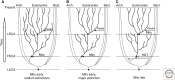
Anton van Leeuwenhoek's startling microscopic observations in the 1600s first stimulated fascination with the way that cells use cilia to generate currents and to swim in a fluid environment. Research in recent decades has yielded deep knowledge about the mechanical and biochemical nature of these organelles but only opened a greater fascination about how such beautifully intricate and multifunctional structures arose during evolution. Answers to this evolutionary puzzle are not only sought to satisfy basic curiosity, but also, as stated so eloquently by Dobzhansky (Am Zool 4: 443 [1964]), because "nothing in biology makes sense except in the light of evolution." Here I attempt to summarize current knowledge of what ciliary organelles of the last eukaryotic common ancestor (LECA) were like, explore the ways in which cilia have evolved since that time, and speculate on the selective processes that might have generated these organelles during early eukaryotic evolution.
Copyright © 2017 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
