Transcriptomic analysis of the highly efficient oil-degrading bacterium Acinetobacter venetianus RAG-1 reveals genes important in dodecane uptake and utilization
- PMID: 27664055
- PMCID: PMC5074533
- DOI: 10.1093/femsle/fnw224
Transcriptomic analysis of the highly efficient oil-degrading bacterium Acinetobacter venetianus RAG-1 reveals genes important in dodecane uptake and utilization
Abstract
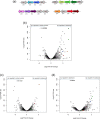
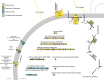
The hydrocarbonoclastic bacterium Acinetobacter venetianus RAG-1 has attracted substantial attention due to its powerful oil-degrading capabilities and its potential to play an important ecological role in the cleanup of alkanes. In this study, we compare the transcriptome of the strain RAG-1 grown in dodecane, the corresponding alkanol (dodecanol), and sodium acetate for the characterization of genes involved in dodecane uptake and utilization. Comparison of the transcriptional responses of RAG-1 grown on dodecane led to the identification of 1074 genes that were differentially expressed relative to sodium acetate. Of these, 622 genes were upregulated when grown in dodecane. The highly upregulated genes were involved in alkane catabolism, along with stress response. Our data suggest AlkMb to be primarily involved in dodecane oxidation. Transcriptional response of RAG-1 grown on dodecane relative to dodecanol also led to the identification of permease, outer membrane protein and thin fimbriae coding genes potentially involved in dodecane uptake. This study provides the first model for key genes involved in alkane uptake and metabolism in A. venetianus RAG-1.
Keywords: Acinetobacter venetianus RAG-1 ATCC 31012; alkane hydroxylase; alkane monooxygenase; alkane uptake; dodecane; transcriptomic.
© FEMS 2016.
Figures
Similar articles
-
A Novel FadL Homolog, AltL, Mediates Transport of Long-Chain Alkanes and Fatty Acids in Acinetobacter venetianus RAG-1.Appl Environ Microbiol. 2022 Oct 26;88(20):e0129422. doi: 10.1128/aem.01294-22. Epub 2022 Sep 28. Appl Environ Microbiol. 2022. PMID: 36169310 Free PMC article.
-
Regulation mechanism of the long-chain n-alkane monooxygenase gene almA in Acinetobacter venetianus RAG-1.Appl Environ Microbiol. 2025 Jan 31;91(1):e0205024. doi: 10.1128/aem.02050-24. Epub 2024 Dec 26. Appl Environ Microbiol. 2025. PMID: 39723816 Free PMC article.
-
Utilization of n-alkanes by a newly isolated strain of Acinetobacter venetianus: the role of two AlkB-type alkane hydroxylases.Appl Microbiol Biotechnol. 2006 Sep;72(2):353-60. doi: 10.1007/s00253-005-0262-9. Epub 2006 Mar 7. Appl Microbiol Biotechnol. 2006. PMID: 16520925
-
Modifications and applications of the Acinetobacter venetianus RAG-1 exopolysaccharide, the emulsan complex and its components.Appl Microbiol Biotechnol. 2008 Nov;81(2):201-10. doi: 10.1007/s00253-008-1664-2. Epub 2008 Sep 16. Appl Microbiol Biotechnol. 2008. PMID: 18795287 Review.
-
Alkane hydroxylases involved in microbial alkane degradation.Appl Microbiol Biotechnol. 2007 Feb;74(1):13-21. doi: 10.1007/s00253-006-0748-0. Epub 2007 Jan 11. Appl Microbiol Biotechnol. 2007. PMID: 17216462 Review.
Cited by
-
Potential of Indigenous Strains Isolated from the Wastewater Treatment Plant of a Crude Oil Refinery.Microorganisms. 2023 Mar 15;11(3):752. doi: 10.3390/microorganisms11030752. Microorganisms. 2023. PMID: 36985325 Free PMC article.
-
Characterization and Transcriptome Analysis of a Long-Chain n-Alkane-Degrading Strain Acinetobacter pittii SW-1.Int J Environ Res Public Health. 2021 Jun 11;18(12):6365. doi: 10.3390/ijerph18126365. Int J Environ Res Public Health. 2021. PMID: 34208299 Free PMC article.
References
-
- Alibrahim M, Shlewit H. Solvent extraction of uranium (VI) by tributyl phosphate/dodecane from nitric acid medium. Period Polytech Chem. 2007;51:57–60.
-
- Andersson MM, Breccia JD, Hatti-Kaul R. Stabilizing effect of chemical additives against oxidation of lactate dehydrogenase. Biotechnol Appl Biochem. 2000;32:145–153. - PubMed
-
- Baumgaertner F, Finsterwalder L. On the transfer mechanism of uranium(VI) and plutonium(IV) nitrate in the system nitric acid-water/tributylphosphate-dodecane. J Phys Chem. 1970;74:108–12.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

