Modeling Small Noncanonical RNA Motifs with the Rosetta FARFAR Server
- PMID: 27665600
- PMCID: PMC7946433
- DOI: 10.1007/978-1-4939-6433-8_12
Modeling Small Noncanonical RNA Motifs with the Rosetta FARFAR Server
Abstract
Noncanonical RNA motifs help define the vast complexity of RNA structure and function, and in many cases, these loops and junctions are on the order of only ten nucleotides in size. Unfortunately, despite their small size, there is no reliable method to determine the ensemble of lowest energy structures of junctions and loops at atomic accuracy. This chapter outlines straightforward protocols using a webserver for Rosetta Fragment Assembly of RNA with Full Atom Refinement (FARFAR) ( http://rosie.rosettacommons.org/rna_denovo/submit ) to model the 3D structure of small noncanonical RNA motifs for use in visualizing motifs and for further refinement or filtering with experimental data such as NMR chemical shifts.
Keywords: RNA 3D structure prediction; RNA Motifs.
Figures
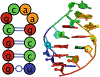




Similar articles
-
Atomic accuracy in predicting and designing noncanonical RNA structure.Nat Methods. 2010 Apr;7(4):291-4. doi: 10.1038/nmeth.1433. Epub 2010 Feb 28. Nat Methods. 2010. PMID: 20190761 Free PMC article.
-
Modeling complex RNA tertiary folds with Rosetta.Methods Enzymol. 2015;553:35-64. doi: 10.1016/bs.mie.2014.10.051. Epub 2015 Feb 12. Methods Enzymol. 2015. PMID: 25726460
-
Secondary Structure Prediction of Single Sequences Using RNAstructure.Methods Mol Biol. 2016;1490:15-34. doi: 10.1007/978-1-4939-6433-8_2. Methods Mol Biol. 2016. PMID: 27665590
-
The RNA 3D Motif Atlas: Computational methods for extraction, organization and evaluation of RNA motifs.Methods. 2016 Jul 1;103:99-119. doi: 10.1016/j.ymeth.2016.04.025. Epub 2016 Apr 25. Methods. 2016. PMID: 27125735 Free PMC article. Review.
-
Graph Theoretical Methods and Workflows for Searching and Annotation of RNA Tertiary Base Motifs and Substructures.Int J Mol Sci. 2021 Aug 9;22(16):8553. doi: 10.3390/ijms22168553. Int J Mol Sci. 2021. PMID: 34445259 Free PMC article. Review.
Cited by
-
F-RAG: Generating Atomic Coordinates from RNA Graphs by Fragment Assembly.J Mol Biol. 2017 Nov 24;429(23):3587-3605. doi: 10.1016/j.jmb.2017.09.017. Epub 2017 Oct 5. J Mol Biol. 2017. PMID: 28988954 Free PMC article.
-
Repurposing of thermally stable nucleic-acid aptamers for targeting tetrodotoxin (TTX).Comput Struct Biotechnol J. 2022 Apr 28;20:2134-2142. doi: 10.1016/j.csbj.2022.04.033. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35832627 Free PMC article.
-
Assessing Exhaustiveness of Stochastic Sampling for Integrative Modeling of Macromolecular Structures.Biophys J. 2017 Dec 5;113(11):2344-2353. doi: 10.1016/j.bpj.2017.10.005. Biophys J. 2017. PMID: 29211988 Free PMC article.
-
Nuclear Magnetic Resonance Structure of an 8 × 8 Nucleotide RNA Internal Loop Flanked on Each Side by Three Watson-Crick Pairs and Comparison to Three-Dimensional Predictions.Biochemistry. 2017 Jul 25;56(29):3733-3744. doi: 10.1021/acs.biochem.7b00201. Epub 2017 Jul 12. Biochemistry. 2017. PMID: 28700212 Free PMC article.
-
Advances in RNA 3D Structure Modeling Using Experimental Data.Front Genet. 2020 Oct 26;11:574485. doi: 10.3389/fgene.2020.574485. eCollection 2020. Front Genet. 2020. PMID: 33193680 Free PMC article. Review.
References
-
- Cech TR, Steitz JA (2014) The noncoding RNA revolution—trashing old rules to forge new ones. Cell 157(1):77–94 - PubMed
-
- Leontis NB, Westhof E (2003) Analysis of RNA motifs. Curr Opin Struct Biol 13(3):300–308 - PubMed
-
- Hendrix DK, Brenner SE, Holbrook SR (2006) RNA structural motifs: building blocks of a modular biomolecule. Q Rev Biophys 38(03):221 - PubMed
-
- Moore PB (1999) Structural motifs in RNA. Annu Rev Biochem 68(1):287–300 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

