Paraspeckles modulate the intranuclear distribution of paraspeckle-associated Ctn RNA
- PMID: 27665741
- PMCID: PMC5036046
- DOI: 10.1038/srep34043
Paraspeckles modulate the intranuclear distribution of paraspeckle-associated Ctn RNA
Abstract
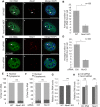
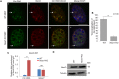
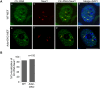
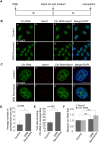
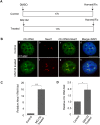
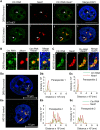
Paraspeckles are sub-nuclear domains that are nucleated by long noncoding RNA Neat1. While interaction of protein components of paraspeckles and Neat1 is understood, there is limited information on the interaction of non-structural RNA components with paraspeckles. Here, by varying paraspeckle number and size, we investigate how paraspeckles influence the nuclear organization of their non-structural RNA component Ctn RNA. Our results show that Ctn RNA remains nuclear-retained in the absence of intact paraspeckles, suggesting that they do not regulate nuclear retention of Ctn RNA. In the absence of Neat1, Ctn RNA continues to interact with paraspeckle protein NonO to form residual nuclear foci. In addition, in the absence of Neat1-nucleated paraspeckles, a subset of Ctn RNA localizes to the perinucleolar regions. Concomitant with increase in number of paraspeckles, transcriptional reactivation resulted in increased number of paraspeckle-localized Ctn RNA foci. Similar to Neat1, proteasome inhibition altered the localization of Ctn RNA, where it formed enlarged paraspeckle-like foci. Super-resolution structured illumination microscopic analyses revealed that in paraspeckles, Ctn RNA partially co-localized with Neat1, and displayed a more heterogeneous intra-paraspeckle localization. Collectively, these results show that while paraspeckles do not influence nuclear retention of Ctn RNA, they modulate its intranuclear compartmentalization.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

