Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration
- PMID: 27666519
- PMCID: PMC5052682
- DOI: 10.1038/ncomms12910
Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration
Abstract
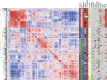
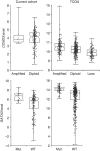
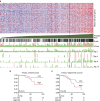
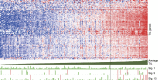
A recent comprehensive whole genome analysis of a large breast cancer cohort was used to link known and novel drivers and substitution signatures to the transcriptome of 266 cases. Here, we validate that subtype-specific aberrations show concordant expression changes for, for example, TP53, PIK3CA, PTEN, CCND1 and CDH1. We find that CCND3 expression levels do not correlate with amplification, while increased GATA3 expression in mutant GATA3 cancers suggests GATA3 is an oncogene. In luminal cases the total number of substitutions, irrespective of type, associates with cell cycle gene expression and adverse outcome, whereas the number of mutations of signatures 3 and 13 associates with immune-response specific gene expression, increased numbers of tumour-infiltrating lymphocytes and better outcome. Thus, while earlier reports imply that the sheer number of somatic aberrations could trigger an immune-response, our data suggests that substitutions of a particular type are more effective in doing so than others.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

