Heritable components of the human fecal microbiome are associated with visceral fat
- PMID: 27666579
- PMCID: PMC5036307
- DOI: 10.1186/s13059-016-1052-7
Heritable components of the human fecal microbiome are associated with visceral fat
Abstract
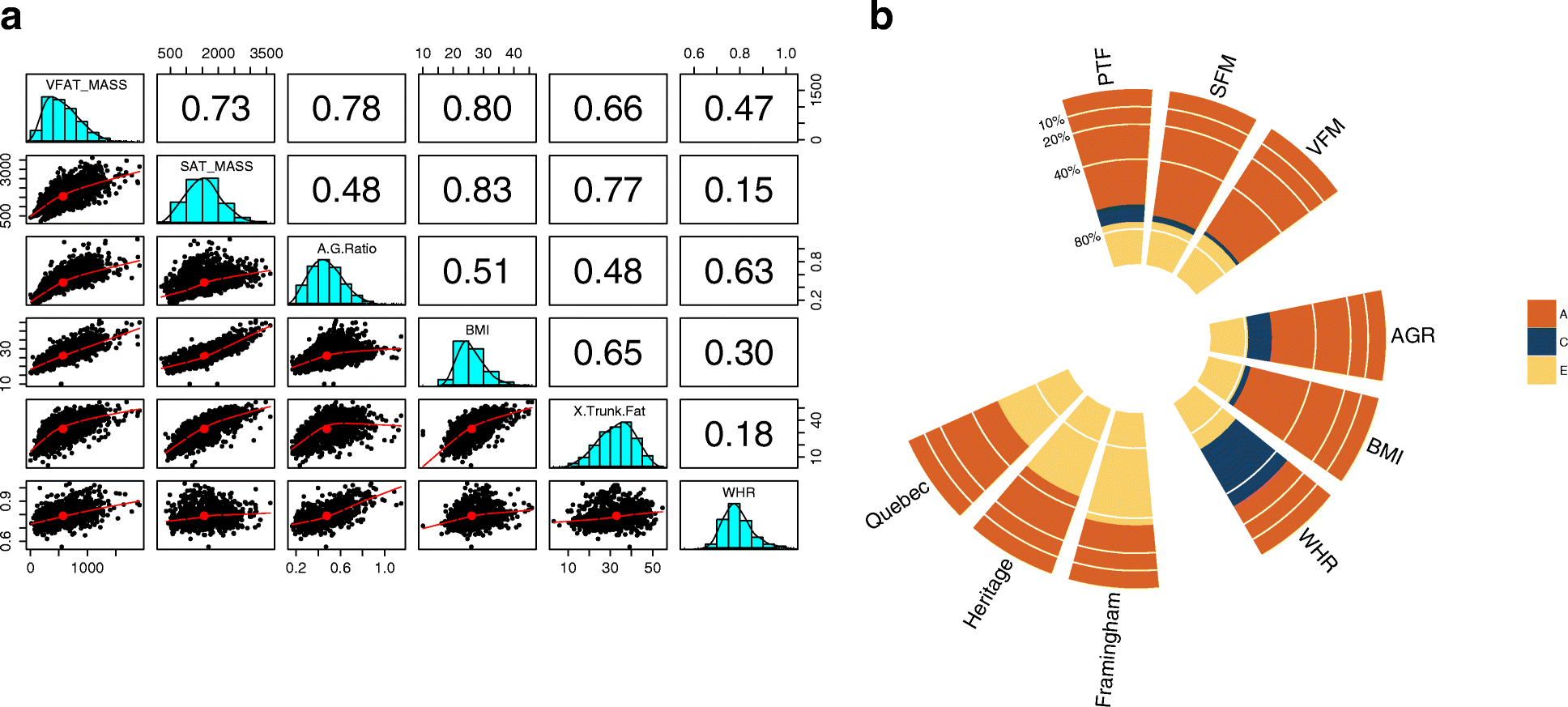
Background: Variation in the human fecal microbiota has previously been associated with body mass index (BMI). Although obesity is a global health burden, the accumulation of abdominal visceral fat is the specific cardio-metabolic disease risk factor. Here, we explore links between the fecal microbiota and abdominal adiposity using body composition as measured by dual-energy X-ray absorptiometry in a large sample of twins from the TwinsUK cohort, comparing fecal 16S rRNA diversity profiles with six adiposity measures.
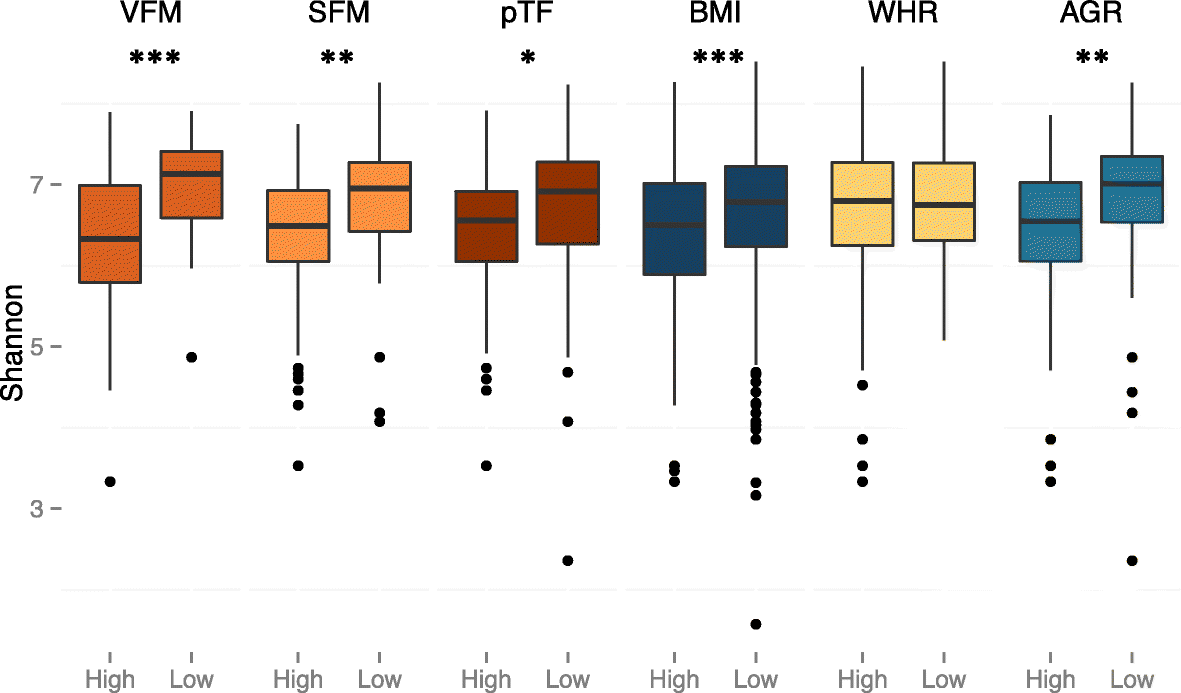
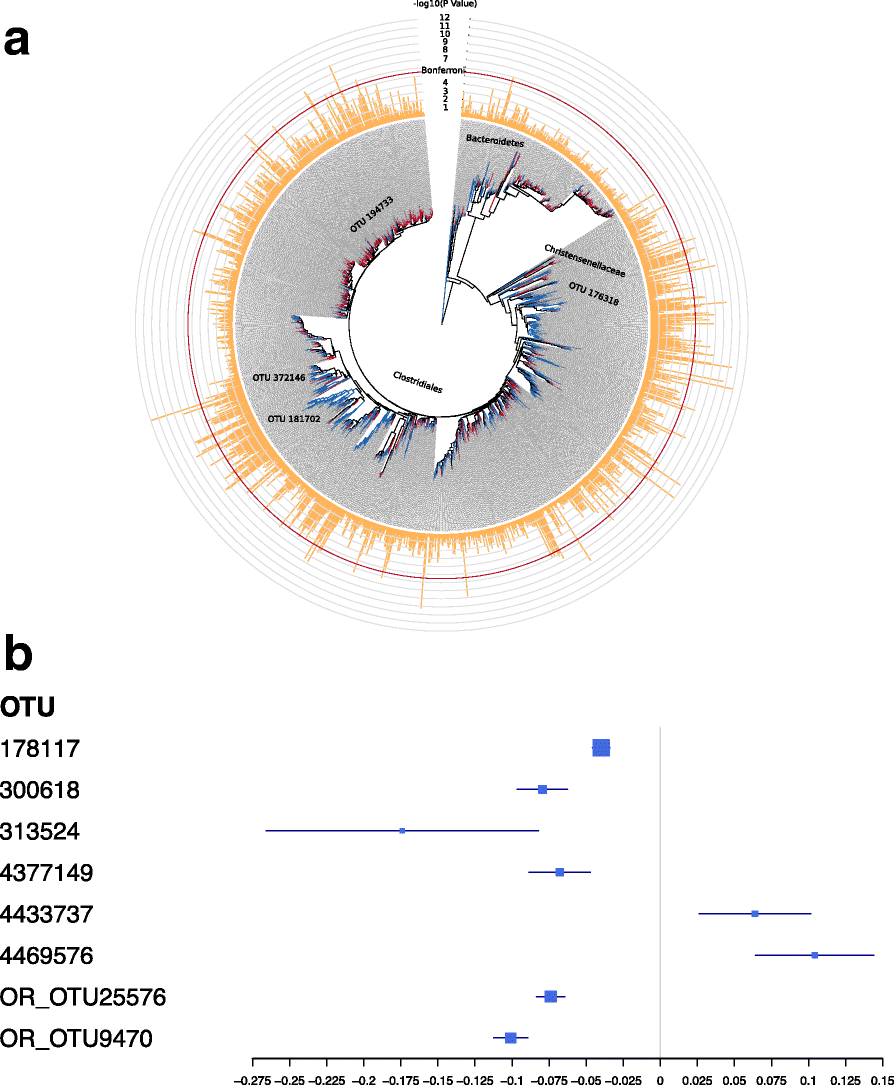
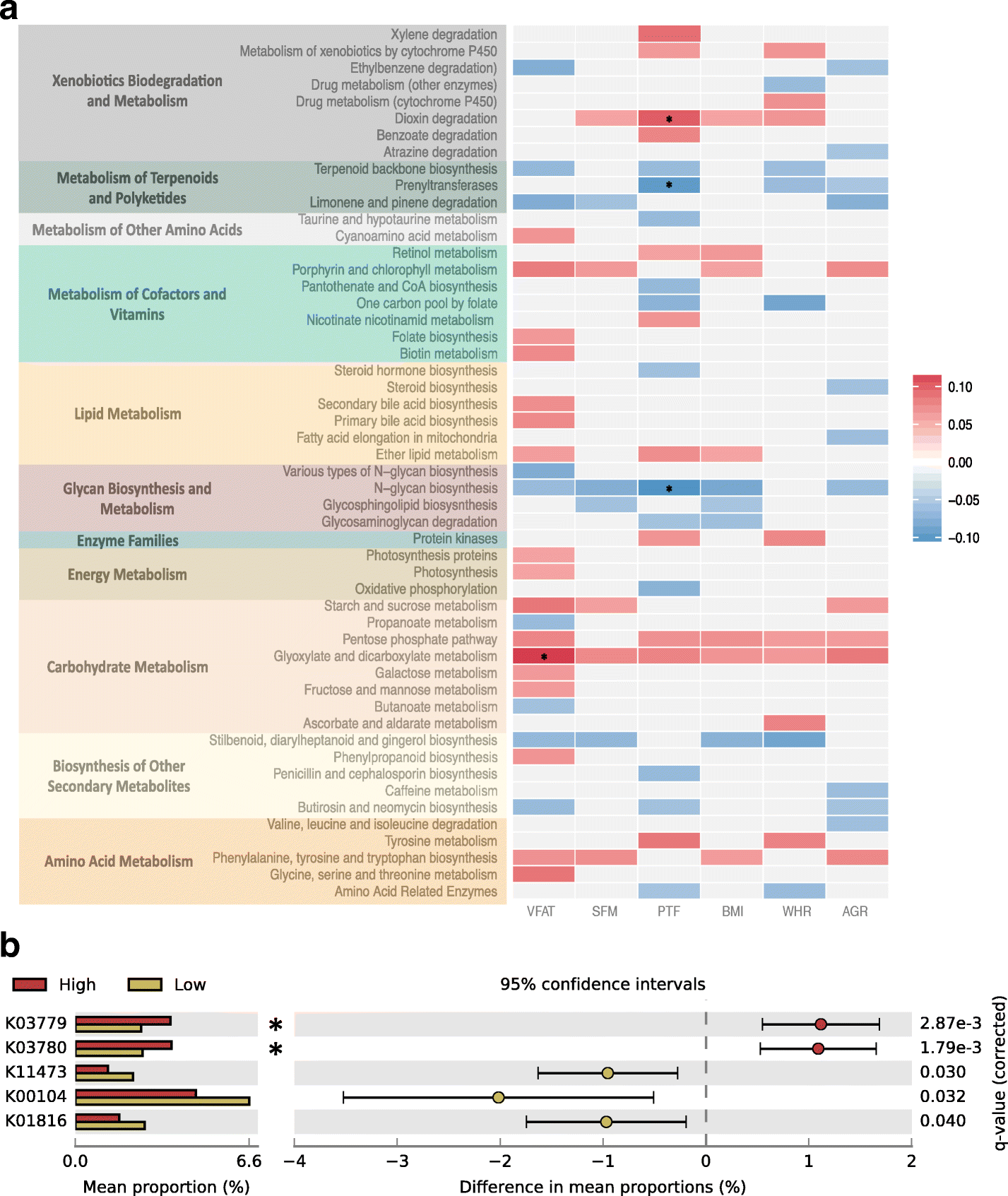
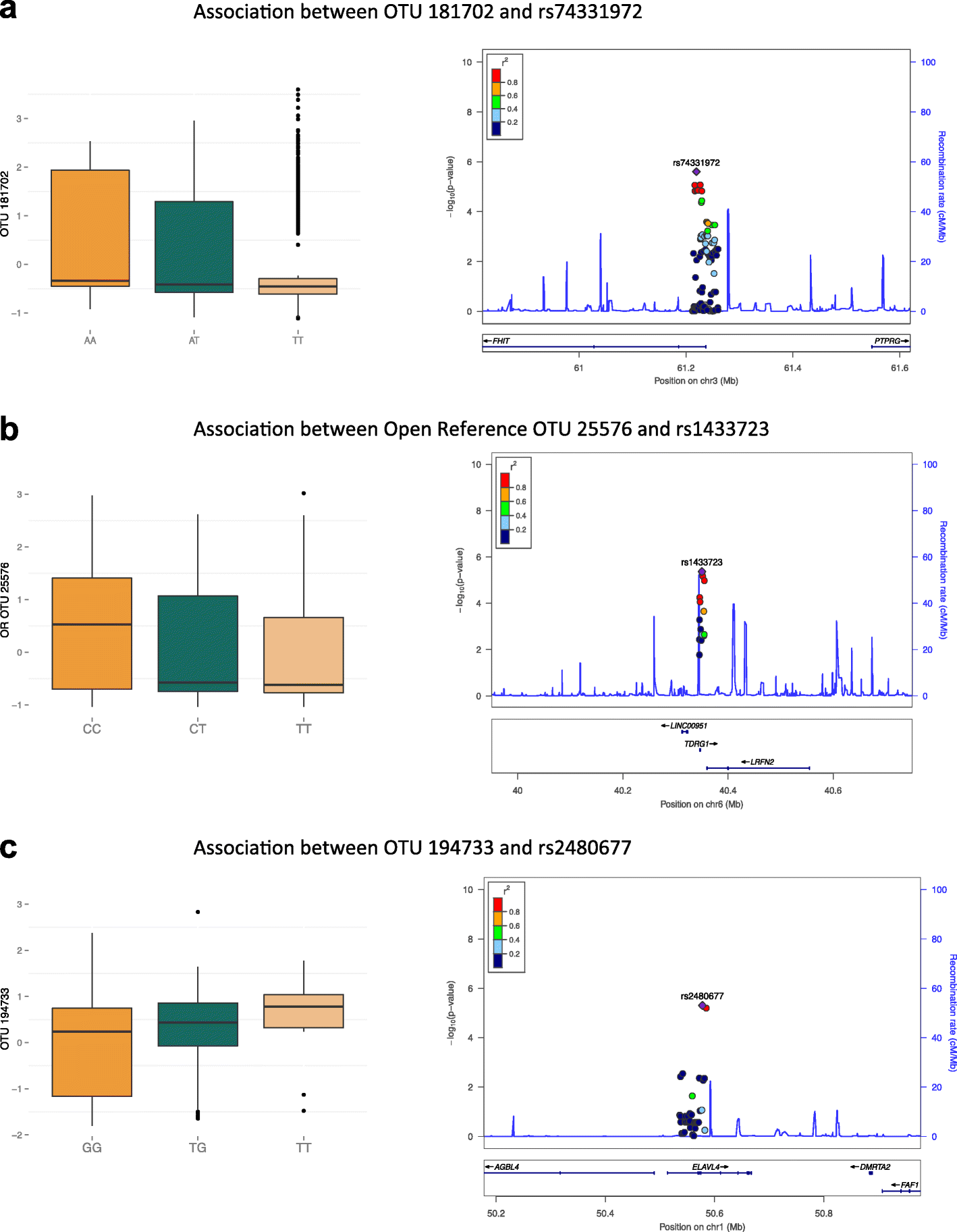
Results: We profile six adiposity measures in 3666 twins and estimate their heritability, finding novel evidence for strong genetic effects underlying visceral fat and android/gynoid ratio. We confirm the association of lower diversity of the fecal microbiome with obesity and adiposity measures, and then compare the association between fecal microbial composition and the adiposity phenotypes in a discovery subsample of twins. We identify associations between the relative abundances of fecal microbial operational taxonomic units (OTUs) and abdominal adiposity measures. Most of these results involve visceral fat associations, with the strongest associations between visceral fat and Oscillospira members. Using BMI as a surrogate phenotype, we pursue replication in independent samples from three population-based cohorts including American Gut, Flemish Gut Flora Project and the extended TwinsUK cohort. Meta-analyses across the replication samples indicate that 8 OTUs replicate at a stringent threshold across all cohorts, while 49 OTUs achieve nominal significance in at least one replication sample. Heritability analysis of the adiposity-associated microbial OTUs prompted us to assess host genetic-microbe interactions at obesity-associated human candidate loci. We observe significant associations of adiposity-OTU abundances with host genetic variants in the FHIT, TDRG1 and ELAVL4 genes, suggesting a potential role for host genes to mediate the link between the fecal microbiome and obesity.
Conclusions: Our results provide novel insights into the role of the fecal microbiota in cardio-metabolic disease with clear potential for prevention and novel therapies.
Keywords: Fecal microbiome; Genetic association; Heritability; Obesity; Twins; Visceral fat.
Figures
Similar articles
-
Heritable components of the human fecal microbiome are associated with visceral fat.Gut Microbes. 2018 Jan 2;9(1):61-67. doi: 10.1080/19490976.2017.1356556. Epub 2017 Aug 11. Gut Microbes. 2018. PMID: 28767316 Free PMC article.
-
The relationship between DXA-based and anthropometric measures of visceral fat and morbidity in women.BMC Cardiovasc Disord. 2013 Apr 3;13:25. doi: 10.1186/1471-2261-13-25. BMC Cardiovasc Disord. 2013. PMID: 23552273 Free PMC article.
-
The associations of the gut microbiome composition and short-chain fatty acid concentrations with body fat distribution in children.Clin Nutr. 2021 May;40(5):3379-3390. doi: 10.1016/j.clnu.2020.11.014. Epub 2020 Nov 20. Clin Nutr. 2021. PMID: 33277072
-
Visceral adiposity and inflammatory bowel disease.Int J Colorectal Dis. 2021 Nov;36(11):2305-2319. doi: 10.1007/s00384-021-03968-w. Epub 2021 Jun 9. Int J Colorectal Dis. 2021. PMID: 34104989 Review.
-
Beverage Consumption and Growth, Size, Body Composition, and Risk of Overweight and Obesity: A Systematic Review [Internet].Alexandria (VA): USDA Nutrition Evidence Systematic Review; 2020 Jul. Alexandria (VA): USDA Nutrition Evidence Systematic Review; 2020 Jul. PMID: 35349233 Free Books & Documents. Review.
Cited by
-
Toxicity, Pharmacokinetics, and Gut Microbiome of Oral Administration of Sesterterpene MHO7 Derived from a Marine Fungus.Mar Drugs. 2019 Nov 26;17(12):667. doi: 10.3390/md17120667. Mar Drugs. 2019. PMID: 31779201 Free PMC article.
-
Effects of high fructose corn syrup on intestinal microbiota structure and obesity in mice.NPJ Sci Food. 2022 Mar 2;6(1):17. doi: 10.1038/s41538-022-00133-7. NPJ Sci Food. 2022. PMID: 35236837 Free PMC article.
-
Darwinian selection of host and bacteria supports emergence of Lamarckian-like adaptation of the system as a whole.Biol Direct. 2018 Oct 26;13(1):24. doi: 10.1186/s13062-018-0224-7. Biol Direct. 2018. PMID: 30621755 Free PMC article.
-
Integrating tumor genomics into studies of the microbiome in colorectal cancer.Gut Microbes. 2019;10(4):547-552. doi: 10.1080/19490976.2018.1549421. Epub 2018 Dec 17. Gut Microbes. 2019. PMID: 30556775 Free PMC article.
-
Preventive Effect of Spontaneous Physical Activity on the Gut-Adipose Tissue in a Mouse Model That Mimics Crohn's Disease Susceptibility.Cells. 2019 Jan 9;8(1):33. doi: 10.3390/cells8010033. Cells. 2019. PMID: 30634469 Free PMC article.
References
-
- Allison DB, Kaprio J, Korkeila M, Koskenvuo M, Neale MC, Hayakawa K. The heritability of body mass index among an international sample of monozygotic twins reared apart. Int J Obes. 1996;20:501–6. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

