Balancing the Robustness and Efficiency of Odor Representations during Learning
- PMID: 27667005
- PMCID: PMC5061050
- DOI: 10.1016/j.neuron.2016.09.004
Balancing the Robustness and Efficiency of Odor Representations during Learning
Abstract
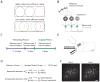
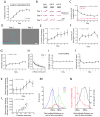
For reliable stimulus identification, sensory codes have to be robust by including redundancy to combat noise, but redundancy sacrifices coding efficiency. To address how experience affects the balance between the robustness and efficiency of sensory codes, we probed odor representations in the mouse olfactory bulb during learning over a week, using longitudinal two-photon calcium imaging. When mice learned to discriminate between two dissimilar odorants, responses of mitral cell ensembles to the two odorants gradually became less discrete, increasing the efficiency. In contrast, when mice learned to discriminate between two very similar odorants, the initially overlapping representations of the two odorants became progressively decorrelated, enhancing the robustness. Qualitatively similar changes were observed when the same odorants were experienced passively, a condition that would induce implicit perceptual learning. These results suggest that experience adjusts odor representations to balance the robustness and efficiency depending on the similarity of the experienced odorants.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
-
- Abraham NM, Spors H, Carleton A, Margrie TW, Kuner T, Schaefer AT. Maintaining accuracy at the expense of speed: Stimulus similarity defines odor discrimination time in mice. Neuron. 2004;44:865–876. - PubMed
-
- Alonso M, Lepousez G, Wagner S, Bardy C, Gabellec MM, Torquet N, Lledo PM. Activation of adult-born neurons facilitates learning and memory. Nat Neurosci. 2012;15:897–904. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

