Filling the Void: Proximity-Based Labeling of Proteins in Living Cells
- PMID: 27667171
- PMCID: PMC5077660
- DOI: 10.1016/j.tcb.2016.09.004
Filling the Void: Proximity-Based Labeling of Proteins in Living Cells
Abstract
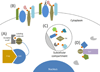
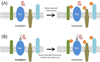
There are inherent limitations with traditional methods to study protein behavior or to determine the constituency of proteins in discrete subcellular compartments. In response to these limitations, several methods have recently been developed that use proximity-dependent labeling. By fusing proteins to enzymes that generate reactive molecules, most commonly biotin, proximate proteins are covalently labeled to enable their isolation and identification. In this review we describe current methods for proximity-dependent labeling in living cells and discuss their applications and future use in the study of protein behavior.
Keywords: APEX; BioID; protein–protein interactions; proteomics; proximity-dependent labeling; subcellular proteome.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

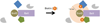

References
-
- Kulyyassov A, et al. PUB-MS: a mass spectrometry-based method to monitor protein-protein proximity in vivo. J Proteome Res. 2011;10(10):4416–4427. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

