Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd
- PMID: 27667448
- PMCID: PMC5052684
- DOI: 10.1038/ncomms12846
Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd
Abstract
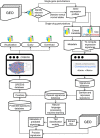
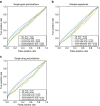
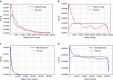
Gene expression data are accumulating exponentially in public repositories. Reanalysis and integration of themed collections from these studies may provide new insights, but requires further human curation. Here we report a crowdsourcing project to annotate and reanalyse a large number of gene expression profiles from Gene Expression Omnibus (GEO). Through a massive open online course on Coursera, over 70 participants from over 25 countries identify and annotate 2,460 single-gene perturbation signatures, 839 disease versus normal signatures, and 906 drug perturbation signatures. All these signatures are unique and are manually validated for quality. Global analysis of these signatures confirms known associations and identifies novel associations between genes, diseases and drugs. The manually curated signatures are used as a training set to develop classifiers for extracting similar signatures from the entire GEO repository. We develop a web portal to serve these signatures for query, download and visualization.
Figures


References
-
- Fujibuchi W., Kiseleva L., Taniguchi T., Harada H. & Horton P. CellMontage: similar expression profile search server. Bioinformatics 23, 3103–3104 (2007). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

