An efficient protocol for obtaining accurate hydration free energies using quantum chemistry and reweighting from molecular dynamics simulations
- PMID: 27667551
- PMCID: PMC5068830
- DOI: 10.1016/j.bmc.2016.08.031
An efficient protocol for obtaining accurate hydration free energies using quantum chemistry and reweighting from molecular dynamics simulations
Abstract
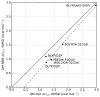
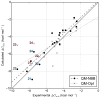
The non-Boltzmann Bennett (NBB) free energy estimator method is applied to 21 molecules from the blind subset of the SAMPL4 challenge. When NBB is applied with the SMD implicit solvent model, and the OLYP/DZP level of quantum chemistry, highly accurate hydration free energy calculations are obtained with respect to experiment (RMSD=0.89kcal·mol-1). Other quantum chemical methods are also tested, and the effects of solvent model, density functional, basis set are explored in this benchmarking study, providing a framework for improvements in calculating hydration free energies. We provide a practical guide for using the best QM-NBB protocols that are consistently more accurate than either pure QM or pure MM alone. In situations where high accuracy hydration free energy predictions are needed, the QM-NBB method with SMD implicit solvent should be the first choice of quantum chemists.
Keywords: Hydration free energy calculations; Implicit solvent; Non-Boltzmann Bennett.
Published by Elsevier Ltd.
Figures


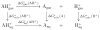

Similar articles
-
Blind prediction of distribution in the SAMPL5 challenge with QM based protomer and pK a corrections.J Comput Aided Mol Des. 2016 Nov;30(11):1087-1100. doi: 10.1007/s10822-016-9955-7. Epub 2016 Sep 19. J Comput Aided Mol Des. 2016. PMID: 27646286
-
Predicting hydration free energies with a hybrid QM/MM approach: an evaluation of implicit and explicit solvation models in SAMPL4.J Comput Aided Mol Des. 2014 Mar;28(3):245-57. doi: 10.1007/s10822-014-9708-4. Epub 2014 Feb 7. J Comput Aided Mol Des. 2014. PMID: 24504703 Free PMC article.
-
Computation of Hydration Free Energies Using the Multiple Environment Single System Quantum Mechanical/Molecular Mechanical Method.J Chem Theory Comput. 2016 Jan 12;12(1):332-44. doi: 10.1021/acs.jctc.5b00874. Epub 2015 Dec 11. J Chem Theory Comput. 2016. PMID: 26613419 Free PMC article.
-
Progress in ab initio QM/MM free-energy simulations of electrostatic energies in proteins: accelerated QM/MM studies of pKa, redox reactions and solvation free energies.J Phys Chem B. 2009 Feb 5;113(5):1253-72. doi: 10.1021/jp8071712. J Phys Chem B. 2009. PMID: 19055405 Free PMC article. Review.
-
QM/MM Calculations on Proteins.Methods Enzymol. 2016;577:119-58. doi: 10.1016/bs.mie.2016.05.014. Epub 2016 Jun 28. Methods Enzymol. 2016. PMID: 27498637 Review.
Cited by
-
Blind prediction of distribution in the SAMPL5 challenge with QM based protomer and pK a corrections.J Comput Aided Mol Des. 2016 Nov;30(11):1087-1100. doi: 10.1007/s10822-016-9955-7. Epub 2016 Sep 19. J Comput Aided Mol Des. 2016. PMID: 27646286
-
Use of Interaction Energies in QM/MM Free Energy Simulations.J Chem Theory Comput. 2019 Aug 13;15(8):4632-4645. doi: 10.1021/acs.jctc.9b00084. Epub 2019 Jul 2. J Chem Theory Comput. 2019. PMID: 31142113 Free PMC article.
-
A Comparison of QM/MM Simulations with and without the Drude Oscillator Model Based on Hydration Free Energies of Simple Solutes.Molecules. 2018 Oct 19;23(10):2695. doi: 10.3390/molecules23102695. Molecules. 2018. PMID: 30347691 Free PMC article.
-
On the faithfulness of molecular mechanics representations of proteins towards quantum-mechanical energy surfaces.Interface Focus. 2020 Dec 6;10(6):20190121. doi: 10.1098/rsfs.2019.0121. Epub 2020 Oct 16. Interface Focus. 2020. PMID: 33184586 Free PMC article.
-
Physics-Based Solubility Prediction for Organic Molecules.Chem Rev. 2025 Aug 13;125(15):7057-7098. doi: 10.1021/acs.chemrev.4c00855. Epub 2025 Jul 29. Chem Rev. 2025. PMID: 40728940 Free PMC article. Review.
References
-
- Nicholls A, Mobley DL, Guthrie JP, Chodera JD, Bayly CI, Cooper MD, Pande VS. Predicting Small-Molecule Solvation Free Energies: An Informal Blind Test for Computational Chemistry. J Med Chem. 2008;51:769–779. - PubMed
-
- Guthrie JP. A Blind Challenge for Computational Solvation Free Energies: Introduction and Overview. J Phys Chem B. 2009;113(14):4501–4507. - PubMed
-
- Marenich AV, Cramer CJ, Truhlar DG. Performance of SM6, SM8, and SMD on the SAMPL1 Test Set for the Prediction of Small-Molecule Solvation Free Energies. J Phys Chem B. 2009a;113(14):4538–4543. - PubMed
-
- Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ. The SAMPL2 blind prediction challenge: introduction and overview. J Comput Aided Mol Des. 2010;24(4):259–279. - PubMed
-
- Klimovich PV, Mobley DL. Predicting hydration free energies using all-atom molecular dynamics simulations and multiple starting conformations. J Comput Aided Mol Des. 2010;24(4):307–316. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

